Punicalagin as an allosteric NSP13 helicase inhibitor potently suppresses SARS-CoV-2 replication in vitro
- PMID: 35985407
- PMCID: PMC9381947
- DOI: 10.1016/j.antiviral.2022.105389
Punicalagin as an allosteric NSP13 helicase inhibitor potently suppresses SARS-CoV-2 replication in vitro
Abstract
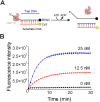
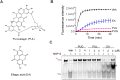
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) helicase NSP13 plays a conserved role in the replication of coronaviruses and has been identified as an ideal target for the development of antiviral drugs against SARS-CoV-2. Here, we identify a novel NSP13 helicase inhibitor punicalagin (PUG) through high-throughput screening. Surface plasmon resonance (SPR)-based analysis and molecular docking calculation reveal that PUG directly binds NSP13 on the interface of domains 1A and 2A, with a KD value of 21.6 nM. Further biochemical and structural analyses suggest that PUG inhibits NSP13 on ATP hydrolysis and prevents it binding to DNA substrates. Finally, the antiviral studies show that PUG effectively suppresses the SARS-CoV-2 replication in A549-ACE2 and Vero cells, with EC50 values of 347 nM and 196 nM, respectively. Our work demonstrates the potential application of PUG in the treatment of coronavirus disease 2019 (COVID-19) and identifies an allosteric inhibition mechanism for future drug design targeting the viral helicases.
Keywords: Helicase; Inhibitor; NSP13; Punicalagin; SARS-CoV-2.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Adaramoye O., Erguen B., Nitzsche B., Hopfner M., Jung K., Rabien A. Punicalagin, a polyphenol from pomegranate fruit, induces growth inhibition and apoptosis in human PC-3 and LNCaP cells. Chem. Biol. Interact. 2017;274:100–106. - PubMed
-
- Cerda B., Ceron J.J., Tomas-Barberan F.A., Espin J.C. Repeated oral administration of high doses of the pomegranate ellagitannin punicalagin to rats for 37 days is not toxic. J. Agric. Food Chem. 2003;51:3493–3501. - PubMed
-
- Cerda B., Periago P., Espin J.C., Tomas-Barberan F.A. Identification of urolithin a as a metabolite produced by human colon microflora from ellagic acid and related compounds. J. Agric. Food Chem. 2005;53:5571–5576. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

