Stage-specific roles of microbial dysbiosis and metabolic disorders in rheumatoid arthritis
- PMID: 35985811
- PMCID: PMC9664099
- DOI: 10.1136/ard-2022-222871
Stage-specific roles of microbial dysbiosis and metabolic disorders in rheumatoid arthritis
Abstract
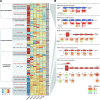
Objective: Rheumatoid arthritis (RA) is a progressive disease including four stages, where gut microbiome is associated with pathogenesis. We aimed to investigate stage-specific roles of microbial dysbiosis and metabolic disorders in RA.
Methods: We investigated stage-based profiles of faecal metagenome and plasma metabolome of 76 individuals with RA grouped into four stages (stages I-IV) according to 2010 RA classification criteria, 19 individuals with osteroarthritis and 27 healthy individuals. To verify bacterial invasion of joint synovial fluid, 16S rRNA gene sequencing, bacterial isolation and scanning electron microscopy were conducted on another validation cohort of 271 patients from four RA stages.
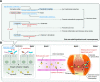
Results: First, depletion of Bacteroides uniformis and Bacteroides plebeius weakened glycosaminoglycan metabolism (p<0.001), continuously hurting articular cartilage across four stages. Second, elevation of Escherichia coli enhanced arginine succinyltransferase pathway in the stage II and stage III (p<0.001), which was correlated with the increase of the rheumatoid factor (p=1.35×10-3) and could induce bone loss. Third, abnormally high levels of methoxyacetic acid (p=1.28×10-8) and cysteine-S-sulfate (p=4.66×10-12) inhibited osteoblasts in the stage II and enhanced osteoclasts in the stage III, respectively, promoting bone erosion. Fourth, continuous increase of gut permeability may induce gut microbial invasion of the joint synovial fluid in the stage IV.
Conclusions: Clinical microbial intervention should consider the RA stage, where microbial dysbiosis and metabolic disorders present distinct patterns and played stage-specific roles. Our work provides a new insight in understanding gut-joint axis from a perspective of stages, which opens up new avenues for RA prognosis and therapy.
Keywords: osteoarthritis; rheumatoid arthritis; rheumatoid factor; synovial fluid.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures


References
-
- Barhum L. What does rheumatoid arthritis progression look like? 2021. Available: https://www.verywellhealth.com/rheumatoid-arthritis-stages-of-progressio...
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

