Vasculogenesis in kidney organoids upon transplantation
- PMID: 35986027
- PMCID: PMC9391397
- DOI: 10.1038/s41536-022-00237-4
Vasculogenesis in kidney organoids upon transplantation
Abstract
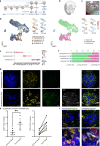
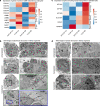
Human induced pluripotent stem cell-derived kidney organoids have potential for disease modeling and to be developed into clinically transplantable auxiliary tissue. However, they lack a functional vasculature, and the sparse endogenous endothelial cells (ECs) are lost upon prolonged culture in vitro, limiting maturation and applicability. Here, we use intracoelomic transplantation in chicken embryos followed by single-cell RNA sequencing and advanced imaging platforms to induce and study vasculogenesis in kidney organoids. We show expansion of human organoid-derived ECs that reorganize into perfused capillaries and form a chimeric vascular network with host-derived blood vessels. Ligand-receptor analysis infers extensive potential interactions of human ECs with perivascular cells upon transplantation, enabling vessel wall stabilization. Perfused glomeruli display maturation and morphogenesis to capillary loop stage. Our findings demonstrate the beneficial effect of vascularization on not only epithelial cell types, but also the mesenchymal compartment, inducing the expansion of ´on target´ perivascular stromal cells, which in turn are required for further maturation and stabilization of the neo-vasculature. The here described vasculogenic capacity of kidney organoids will have to be deployed to achieve meaningful glomerular maturation and kidney morphogenesis in vitro.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
LinkOut - more resources
Full Text Sources

