Opposing roles of CLK SR kinases in controlling HIV-1 gene expression and latency
- PMID: 35986377
- PMCID: PMC9389714
- DOI: 10.1186/s12977-022-00605-4
Opposing roles of CLK SR kinases in controlling HIV-1 gene expression and latency
Abstract
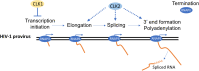
Background: The generation of over 69 spliced HIV-1 mRNAs from one primary transcript by alternative RNA splicing emphasizes the central role that RNA processing plays in HIV-1 replication. Control is mediated in part through the action of host SR proteins whose activity is regulated by multiple SR kinases (CLK1-4, SRPKs).
Methods: Both shRNA depletion and small molecule inhibitors of host SR kinases were used in T cell lines and primary cells to evaluate the role of these factors in the regulation of HIV-1 gene expression. Effects on virus expression were assessed using western blotting, RT-qPCR, and immunofluorescence.
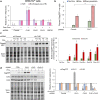
Results: The studies demonstrate that SR kinases play distinct roles; depletion of CLK1 enhanced HIV-1 gene expression, reduction of CLK2 or SRPK1 suppressed it, whereas CLK3 depletion had a modest impact. The opposing effects of CLK1 vs. CLK2 depletion were due to action at distinct steps; reduction of CLK1 increased HIV-1 promoter activity while depletion of CLK2 affected steps after transcript initiation. Reduced CLK1 expression also enhanced the response to several latency reversing agents, in part, by increasing the frequency of responding cells, consistent with a role in regulating provirus latency. To determine whether small molecule modulation of SR kinase function could be used to control HIV-1 replication, we screened a GSK library of protein kinase inhibitors (PKIS) and identified several pyrazolo[1,5-b] pyridazine derivatives that suppress HIV-1 gene expression/replication with an EC50 ~ 50 nM. The compounds suppressed HIV-1 protein and viral RNA accumulation with minimal impact on cell viability, inhibiting CLK1 and CLK2 but not CLK3 function, thereby selectively altering the abundance of individual CLK and SR proteins in cells.
Conclusions: These findings demonstrate the unique roles played by individual SR kinases in regulating HIV-1 gene expression, validating the targeting of these functions to either enhance latency reversal, essential for "Kick-and-Kill" strategies, or to silence HIV protein expression for "Block-and-Lock" strategies.
Keywords: HIV-1; Latency; RNA processing; SR kinases.
Plain language summary
Identifying cellular factors that regulate HIV-1 RNA processing provides important insights into novel strategies to control this infection. Different members of the SR kinase family have distinct roles in regulating virus expression because they affect distinct steps of transcription/RNA processing. We identify inhibitors of these kinases that suppress HIV-1 gene expression and replication in multiple assay systems at nanomolar concentrations with limited or no cytotoxicity. Our results highlight the therapeutic potential of targeting the post-integration stage of the HIV-1 lifecycle to selectively enhance or reverse provirus latency. A greater understanding of the molecular mechanisms underlying the effects observed will facilitate the development of more targeted approaches to modulate HIV-1 latency on the path toward a “functional” cure for this infection.
© 2022. The Author(s).
Conflict of interest statement
The authors have no competing interests as defined by BMC, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
Figures






Similar articles
-
Differential effect of CLK SR Kinases on HIV-1 gene expression: potential novel targets for therapy.Retrovirology. 2011 Jun 17;8:47. doi: 10.1186/1742-4690-8-47. Retrovirology. 2011. PMID: 21682887 Free PMC article.
-
Posttranscriptional Regulation of HIV-1 Gene Expression during Replication and Reactivation from Latency by Nuclear Matrix Protein MATR3.mBio. 2018 Nov 13;9(6):e02158-18. doi: 10.1128/mBio.02158-18. mBio. 2018. PMID: 30425153 Free PMC article.
-
Human CDC2-like kinase 1 (CLK1): a novel target for Alzheimer's disease.Curr Drug Targets. 2014 May;15(5):539-50. doi: 10.2174/1389450115666140226112321. Curr Drug Targets. 2014. PMID: 24568585 Review.
-
Regulation of influenza A virus mRNA splicing by CLK1.Antiviral Res. 2019 Aug;168:187-196. doi: 10.1016/j.antiviral.2019.06.003. Epub 2019 Jun 6. Antiviral Res. 2019. PMID: 31176694
-
HIV-1 Latency and Viral Reservoirs: Existing Reversal Approaches and Potential Technologies, Targets, and Pathways Involved in HIV Latency Studies.Cells. 2021 Feb 23;10(2):475. doi: 10.3390/cells10020475. Cells. 2021. PMID: 33672138 Free PMC article. Review.
Cited by
-
Exploiting the Achilles' Heel of Viral RNA Processing to Develop Novel Antivirals.Viruses. 2024 Dec 31;17(1):54. doi: 10.3390/v17010054. Viruses. 2024. PMID: 39861843 Free PMC article.
-
Viral Modulation of Host Splicing.Annu Rev Virol. 2025 Apr 25:10.1146/annurev-virology-092623-102539. doi: 10.1146/annurev-virology-092623-102539. Online ahead of print. Annu Rev Virol. 2025. PMID: 40279630 Review.
-
The anticancer potential of the CLK kinases inhibitors 1C8 and GPS167 revealed by their impact on the epithelial-mesenchymal transition and the antiviral immune response.Oncotarget. 2024 May 16;15:313-325. doi: 10.18632/oncotarget.28585. Oncotarget. 2024. PMID: 38753413 Free PMC article.
-
Serine-arginine protein kinases and their targets in viral infection and their inhibition.Cell Mol Life Sci. 2023 May 17;80(6):153. doi: 10.1007/s00018-023-04808-6. Cell Mol Life Sci. 2023. PMID: 37198350 Free PMC article. Review.
-
Endogenous Retroviruses Unveiled: A Comprehensive Review of Inflammatory Signaling/Senescence-Related Pathways and Therapeutic Strategies.Aging Dis. 2024 May 14;16(2):738-756. doi: 10.14336/AD.2024.0123-1. Aging Dis. 2024. PMID: 38916727 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

