Calf rumen microbiome from birth to weaning and shared microbial properties to the maternal rumen microbiome
- PMID: 35986918
- PMCID: PMC9576027
- DOI: 10.1093/jas/skac264
Calf rumen microbiome from birth to weaning and shared microbial properties to the maternal rumen microbiome
Abstract
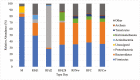
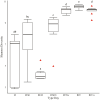
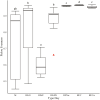
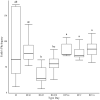
Optimization of host performance in cattle may be achieved through programming of the rumen microbiome. Thus, understanding maternal influences on the development of the calf rumen microbiome is critical. We hypothesized that there exists a shared microbial profile between the cow and calf rumen microbiomes from birth through weaning. Specifically, our objective was to relate the calf's meconium and rumen fluid microbiomes in early life to that of the cow rumen fluid prior to parturition and at weaning. Rumen fluid was collected from multiparous Angus crossbred cows (n = 10) prior to parturition and at weaning. Immediately following the parturition, meconium and rumen fluid were collected from the calf. Rumen fluid was collected again from the calf on day 2, day 28, and at weaning. The rumen fluid microbial profile and subsequent volatile fatty acid (VFA) profile were characterized using 16S rRNA sequencing and gas liquid chromatography, respectively. Microbial data was analyzed using QIIME2 and the GLM procedure of SAS was used to analyze the VFA profile. Alpha diversity was similar in the early gut microbiome (meconium, rumen fluid at birth and day 2; q ≥ 0.12) and between the cow and calf at weaning (q ≥ 0.06). Microbial composition, determined by beta diversity, differed in the early rumen microbiome (rumen fluid at birth, day 2, and day 28; q ≤ 0.04), and VFA profiles complimented these results. There were similarities in composition between meconium, rumen fluid at birth, and rumen fluid from the cow at weaning (q ≥ 0.09). These data indicate successive development of the rumen microbiome and stabilization over time. Similarities between meconium and rumen fluid at birth potentially indicates in utero colonization of the calf gastrointestinal tract. Similarities in composition between the early calf rumen microbiome and the cow at weaning prompt an interesting comparison and area for future consideration in terms of identifying at what stage of gestation might colonization begin. Overall, this study provides insight into similarities between the cow and calf microbiomes and may be helpful in developing hypotheses for the pathway of colonization and programming potential in the early gut.
Keywords: meconium; programming; rumen microbiome.
Plain language summary
Developmental programming has highlighted important influences of maternal factors on offspring development. Recent research indicates a programming potential of the rumen microbiome and understanding this role as well as how inoculation occurs may allow beef producers to optimize management practices of gestating cows such that offspring performance is improved via the rumen microbiome. To investigate this, rumen fluid samples were collected from mature cows immediately prior to calving, from their calf immediately after calving with a meconium sample, day 2, and day 28 as well as collected from both dam and calf at weaning. The rumen and meconium microbiome of the newborn calf were similar to each other as well as to the cow rumen microbiome at weaning, although not to the cow rumen microbiome immediately prior to calving. The shared microbiome of the early calf gut highlight a common source of inoculation. The similarities with the cow rumen at weaning could indicate initiation of colonization occurs early in gestation. Results indicate there are shared microbial properties between the cow and calf rumen microbiome. This further supports the opportunity to alter the calf rumen microbiome to improve productivity through the management of the cow during gestation.
© The Author(s) 2022. Published by Oxford University Press on behalf of the American Society of Animal Science. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
Similar articles
-
Maternal gastrointestinal microbiome shapes gut microbial function and resistome of newborns in a cow-to-calf model.Microbiome. 2024 Oct 22;12(1):216. doi: 10.1186/s40168-024-01943-5. Microbiome. 2024. PMID: 39438998 Free PMC article.
-
The effects of including sprouted barley with alfalfa hay in the diet on ruminal health and performance of cow-calf pairs.J Anim Sci. 2024 Jan 3;102:skae074. doi: 10.1093/jas/skae074. J Anim Sci. 2024. PMID: 38502875 Free PMC article.
-
The effect of maternal supply of rumen-protected protein to Holstein Friesian cows during the dry period on the transfer of passive immunity and colostral microbial composition.J Dairy Sci. 2023 Dec;106(12):8723-8745. doi: 10.3168/jds.2023-23266. Epub 2023 Sep 9. J Dairy Sci. 2023. PMID: 37678775
-
Impact of dairy calf management practices on the intestinal tract microbiome pre-weaning.J Med Microbiol. 2025 Jan;74(1). doi: 10.1099/jmm.0.001957. J Med Microbiol. 2025. PMID: 39879083 Review.
-
Development and physiology of the rumen and the lower gut: Targets for improving gut health.J Dairy Sci. 2016 Jun;99(6):4955-4966. doi: 10.3168/jds.2015-10351. Epub 2016 Mar 9. J Dairy Sci. 2016. PMID: 26971143 Review.
Cited by
-
Effect of castration timing and weaning strategy on the taxonomic and functional profile of ruminal bacteria and archaea of beef calves.Anim Microbiome. 2023 Dec 1;5(1):61. doi: 10.1186/s42523-023-00284-2. Anim Microbiome. 2023. PMID: 38041127 Free PMC article.
-
Bovine neonatal microbiome origins: a review of proposed microbial community presence from conception to colostrum.Transl Anim Sci. 2023 May 27;7(1):txad057. doi: 10.1093/tas/txad057. eCollection 2023 Jan. Transl Anim Sci. 2023. PMID: 37334245 Free PMC article.
-
Effects of seabuckthorn pomace on rumen development, intramuscular fatty acids and antioxidant capacity in weaned lambs.Front Vet Sci. 2025 Apr 24;12:1560976. doi: 10.3389/fvets.2025.1560976. eCollection 2025. Front Vet Sci. 2025. PMID: 40343368 Free PMC article.
-
Characterizing the Fermentation of Oat Grass (Avena sativa L.) in the Rumen: Integrating Degradation Kinetics, Ultrastructural Examination with Scanning Electron Microscopy, Surface Enzymatic Activity, and Microbial Community Analysis.Animals (Basel). 2025 Jul 11;15(14):2049. doi: 10.3390/ani15142049. Animals (Basel). 2025. PMID: 40723511 Free PMC article.
-
Whole-body microbiota of newborn calves and their response to prenatal vitamin and mineral supplementation.Front Microbiol. 2023 Jun 26;14:1207601. doi: 10.3389/fmicb.2023.1207601. eCollection 2023. Front Microbiol. 2023. PMID: 37434710 Free PMC article.
References
-
- Amat, S., Holman D. B., Schmidt K., McCarthy K. L., Dorsam S. T., Ward A. K., Borowicz P. P., Reynolds L. P., Caton J. S., Sedivec K. K.,. et al. 2021. Characterization of the microbiota associated with 12-week-old bovine fetuses exposed to divergent in utero nutrition. Front. Microbiol. 12:771832. doi: 10.3389/fmicb.2021.771832. - DOI - PMC - PubMed
-
- Appiah, S. A., Foxx C. L., Langgartner D., Palmer A., Zambrano C. A., Braumüller S., Schaefer E. J., Wachter U., Elam B. L., Radermacher P.,. et al. 2021. Evaluation of the gut microbiome in association with biological signatures of inflammation in murine polytrauma and shock. Sci. Rep. 11:6665. doi: 10.1038/s41598-021-85897-w. - DOI - PMC - PubMed
-
- Ardissone, A. N., de la Cruz D. M., Davis-Richardson A. G., Rechcigl K. T., Li N., Drew J. C., Murgas-Torrazza R., Sharma R., Hudak M. L., Triplett E. W., and Neu J... 2014. Meconium microbiome analysis identifies bacteria correlated with premature birth. PLoS One 9:e90784. doi: 10.1371/journal.pone.0090784. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

