Estimating the population-level prevalence of antimicrobial-resistant enteric bacteria from latrine samples
- PMID: 35987780
- PMCID: PMC9392229
- DOI: 10.1186/s13756-022-01145-4
Estimating the population-level prevalence of antimicrobial-resistant enteric bacteria from latrine samples
Abstract
Background: Logistical and economic barriers hamper community-level surveillance for antimicrobial-resistant bacteria in low-income countries. Latrines are commonly used in these settings and offer a low-cost source of surveillance samples. It is unclear, however, whether antimicrobial resistance prevalence estimates from latrine samples reflect estimates generated from randomly sampled people.
Methods: We compared the prevalence of antimicrobial-resistant enteric bacteria from stool samples of people residing in randomly selected households within Kibera-an informal urban settlement in Kenya-to estimates from latrine samples within the same community. Fecal samples were collected between November 2015 and Jan 2016. Presumptive Escherichia coli isolates were collected from each household stool sample (n = 24) and each latrine sample (n = 48), resulting in 8935 and 8210 isolates, respectively. Isolates were tested for resistance to nine antibiotics using the replica-plating technique. Correlation- and Kolmogorov-Smirnov (K-S) tests were used to compare results.
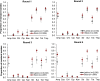
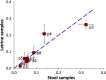
Results: Overall, the prevalence values obtained from latrine samples closely reflected those from stool samples, particularly for low-prevalence (< 15%) resistance phenotypes. Similarly, the distribution of resistance phenotypes was similar between latrine and household samples (r > 0.6; K-S p-values > 0.05).
Conclusions: Although latrine samples did not perfectly estimate household antimicrobial resistance prevalence, they were highly correlated and thus could be employed as low-cost samples to monitor trends in antimicrobial resistance, detect the emergence of new resistance phenotypes and assess the impact of community interventions.
Keywords: Antimicrobial resistance; Latrines; Prevalence estimates; Surveillance.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- World Health Organization, WHO. Antimicrobial Resistance: Global Report on surveillance. Geneva; 2014. https://apps.who.int/iris/handle/10665/112642. Accessed 13 May 2021.
-
- Omulo S, Lofgren ET, Lockwood S, Thumbi SM, Bigogo G, Ouma A, et al. Carriage of antimicrobial-resistant bacteria in a high-density informal settlement in Kenya is associated with environmental risk-factors. Antimicrob Resist Infect Control. 2021;10(1):1–12. doi: 10.1186/s13756-021-00886-y. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

