Genomic targets and selective inhibition of DNA methyltransferase isoforms
- PMID: 35987848
- PMCID: PMC9392947
- DOI: 10.1186/s13148-022-01325-4
Genomic targets and selective inhibition of DNA methyltransferase isoforms
Abstract
Background: DNA methylation in the human genome is established and maintained by DNA methyltransferases (DNMTs). DNMT isoforms show differential expression by cell lineage and during development, but much remains to be elucidated about their shared and unique genomic targets.
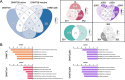
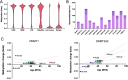
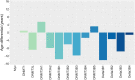
Results: We examined changes in the epigenome following overexpression of 13 DNMT isoforms in HEK293T cells. We observed increased methylation (Δβ > 0.2) at 43,405 CpG sites, with expression of DNMT3A2, DNMTΔ3B4 and DNMTΔ3B2 associated with the greatest impact. De novo methylation occurred primarily within open sea regions and at loci with intermediate methylation levels (β: 0.2-0.6). 53% of differentially methylated loci showed specificity towards a single DNMT subfamily, primarily DNMTΔ3B and DNMT3A and 39% towards a single isoform. These loci were significantly enriched for pathways related to neuronal development (DNMTΔ3B4), calcium homeostasis (DNMTΔ3B3) and ion transport (DNMT3L). Repetitive elements did not display differential sensitivity to overexpressed DNMTs, but hypermethylation of Alu elements was associated with their evolutionary age following overexpression of DNMT3A2, DNMT3B1, DNMT3B2 and DNMT3L. Differential methylation (Δβ > 0.1) was observed at 121 of the 353 loci associated with the Horvath 'epigenetic clock' model of ageing, with 51 showing isoform specificity, and was associated with reduction of epigenetic age by 5-15 years following overexpression of seven isoforms. Finally, we demonstrate the potential for dietary constituents to modify epigenetic marks through isoform-specific inhibition of methylation activity.
Conclusions: Our results provide insight into regions of the genome methylated uniquely by specific DNMT isoforms and demonstrate the potential for dietary intervention to modify the epigenome.
Keywords: DNA methylation; DNA methyltransferases; DNMT isoforms; Dietary constituents; Epigenetic clock; Epigenome; Repetitive elements.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

