A tissue-level phenome-wide network map of colocalized genes and phenotypes in the UK Biobank
- PMID: 35987940
- PMCID: PMC9392744
- DOI: 10.1038/s42003-022-03820-z
A tissue-level phenome-wide network map of colocalized genes and phenotypes in the UK Biobank
Abstract
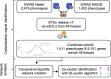
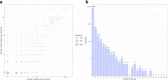
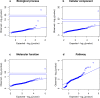
Phenome-wide association studies identified numerous loci associated with traits and diseases. To help interpret these associations, we constructed a phenome-wide network map of colocalized genes and phenotypes. We generated colocalized signals using the Genotype-Tissue Expression data and genome-wide association results in UK Biobank. We identified 9151 colocalized genes for 1411 phenotypes across 48 tissues. Then, we constructed bipartite networks using the colocalized signals in each tissue, and showed that the majority of links were observed in a single tissue. We applied the biLouvain clustering algorithm in each tissue-specific network to identify co-clusters of genes and phenotypes. We observed significant enrichments of these co-clusters with known biological and functional gene classes. Overall, the phenome-wide map provides links between genes, phenotypes and tissues, and can yield biological and clinical discoveries.
© 2022. The Author(s).
Conflict of interest statement
R.D. received grants from AstraZeneca, grants and nonfinancial support from Goldfinch Bio, is a scientific co-founder, equity holder and consultant for Pensieve Health (pending), and is a consultant for Variant Bio, all not related to this work. The remaining authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Grants and funding
- R35 GM124836/GM/NIGMS NIH HHS/United States
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- R01 HL139865/HL/NHLBI NIH HHS/United States
- R01HL139865/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- T32 HL007824/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources

