Post-gastrulation synthetic embryos generated ex utero from mouse naive ESCs
- PMID: 35988542
- PMCID: PMC9439721
- DOI: 10.1016/j.cell.2022.07.028
Post-gastrulation synthetic embryos generated ex utero from mouse naive ESCs
Abstract
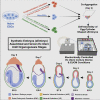
In vitro cultured stem cells with distinct developmental capacities can contribute to embryonic or extraembryonic tissues after microinjection into pre-implantation mammalian embryos. However, whether cultured stem cells can independently give rise to entire gastrulating embryo-like structures with embryonic and extraembryonic compartments remains unknown. Here, we adapt a recently established platform for prolonged ex utero growth of natural embryos to generate mouse post-gastrulation synthetic whole embryo models (sEmbryos), with both embryonic and extraembryonic compartments, starting solely from naive ESCs. This was achieved by co-aggregating non-transduced ESCs, with naive ESCs transiently expressing Cdx2 or Gata4 to promote their priming toward trophectoderm and primitive endoderm lineages, respectively. sEmbryos adequately accomplish gastrulation, advance through key developmental milestones, and develop organ progenitors within complex extraembryonic compartments similar to E8.5 stage mouse embryos. Our findings highlight the plastic potential of naive pluripotent cells to self-organize and functionally reconstitute and model the entire mammalian embryo beyond gastrulation.
Keywords: ESCs; PGCs; embryogenesis; embryoids; ex utero; extra-embryonic Tissues; iPSCs; naive pluripotency; synthetic embryos.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests J.H.H. has submitted patent applications relevant to the findings reported herein and is a chief scientific advisor of Renewal Bio Inc., which has licensed technologies described herein.
Figures









Comment in
-
Mouse embryos grown without eggs or sperm: why, and what's next?Nature. 2022 Sep;609(7926):230-231. doi: 10.1038/d41586-022-02334-2. Nature. 2022. PMID: 36008716 No abstract available.
-
Ex utero synthetic embryos.Nat Methods. 2022 Sep;19(9):1033. doi: 10.1038/s41592-022-01612-1. Nat Methods. 2022. PMID: 36068319 No abstract available.
-
Mouse embryo models built from stem cells take shape in a dish.Nature. 2022 Oct;610(7930):39-40. doi: 10.1038/d41586-022-03075-y. Nature. 2022. PMID: 36192499 Free PMC article.
-
Ensemble of old and new techniques escorts ESCs to bona fide embryo-like structures.Cell Stem Cell. 2022 Oct 6;29(10):1423-1425. doi: 10.1016/j.stem.2022.09.004. Cell Stem Cell. 2022. PMID: 36206728
References
-
- Bayerl J., Ayyash M., Shani T., Manor Y.S., Gafni O., Massarwa R., Kalma Y., Aguilera-Castrejon A., Zerbib M., Amir H., et al. Principles of signaling pathway modulation for enhancing human naive pluripotency induction. Cell Stem Cell. 2021;28:1549–1565.e12. doi: 10.1016/j.stem.2021.04.001. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

