Antibody escape and cryptic cross-domain stabilization in the SARS-CoV-2 Omicron spike protein
- PMID: 35988543
- PMCID: PMC9350683
- DOI: 10.1016/j.chom.2022.07.016
Antibody escape and cryptic cross-domain stabilization in the SARS-CoV-2 Omicron spike protein
Abstract
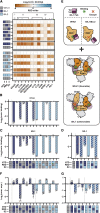
The worldwide spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to the repeated emergence of variants of concern. For the Omicron variant, sub-lineages BA.1 and BA.2, respectively, contain 33 and 29 nonsynonymous and indel spike protein mutations. These amino acid substitutions and indels are implicated in increased transmissibility and enhanced immune evasion. By reverting individual spike mutations of BA.1 or BA.2, we characterize the molecular effects of the Omicron spike mutations on expression, ACE2 receptor affinity, and neutralizing antibody recognition. We identified key mutations enabling escape from neutralizing antibodies at a variety of epitopes. Stabilizing mutations in the N-terminal and S2 domains of the spike protein can compensate for destabilizing mutations in the receptor binding domain, enabling the record number of mutations in Omicron. Our results provide a comprehensive account of the mutational effects in the Omicron spike protein and illustrate previously uncharacterized mechanisms of host evasion.
Keywords: COVID-19; VOCs; cell display; flow cytometry; high throughput; viral glycoprotein.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests D.R.B., A.D.E., and J.D.G. have filed patent applications monoclonal antibodies targeting SARS-CoV-2. K.J., C.-W.C., and I.J.F. have filed patent applications on spike 6p (HexaPro).
Figures



Similar articles
-
AlphaFold2 Modeling and Molecular Dynamics Simulations of the Conformational Ensembles for the SARS-CoV-2 Spike Omicron JN.1, KP.2 and KP.3 Variants: Mutational Profiling of Binding Energetics Reveals Epistatic Drivers of the ACE2 Affinity and Escape Hotspots of Antibody Resistance.Viruses. 2024 Sep 13;16(9):1458. doi: 10.3390/v16091458. Viruses. 2024. PMID: 39339934 Free PMC article.
-
Characterization of Entry Pathways, Species-Specific Angiotensin-Converting Enzyme 2 Residues Determining Entry, and Antibody Neutralization Evasion of Omicron BA.1, BA.1.1, BA.2, and BA.3 Variants.J Virol. 2022 Sep 14;96(17):e0114022. doi: 10.1128/jvi.01140-22. Epub 2022 Aug 24. J Virol. 2022. PMID: 36000843 Free PMC article.
-
BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.Nature. 2022 Aug;608(7923):593-602. doi: 10.1038/s41586-022-04980-y. Epub 2022 Jun 17. Nature. 2022. PMID: 35714668 Free PMC article.
-
Immune evasion of neutralizing antibodies by SARS-CoV-2 Omicron.Cytokine Growth Factor Rev. 2023 Apr;70:13-25. doi: 10.1016/j.cytogfr.2023.03.001. Epub 2023 Mar 5. Cytokine Growth Factor Rev. 2023. PMID: 36948931 Free PMC article. Review.
-
SARS-CoV-2's Variants of Concern: A Brief Characterization.Front Immunol. 2022 Jul 26;13:834098. doi: 10.3389/fimmu.2022.834098. eCollection 2022. Front Immunol. 2022. PMID: 35958548 Free PMC article. Review.
Cited by
-
Evolution of the SARS-CoV-2 Omicron spike.Cell Rep. 2023 Dec 26;42(12):113444. doi: 10.1016/j.celrep.2023.113444. Epub 2023 Nov 18. Cell Rep. 2023. PMID: 37979169 Free PMC article. Review.
-
Balancing stability and function: impact of the surface charge of SARS-CoV-2 Omicron spike protein.Npj Viruses. 2025 Apr 1;3(1):23. doi: 10.1038/s44298-025-00104-1. Npj Viruses. 2025. PMID: 40295844 Free PMC article. Review.
-
SARS-COV-2 Omicron variants conformationally escape a rare quaternary antibody binding mode.Commun Biol. 2023 Dec 11;6(1):1250. doi: 10.1038/s42003-023-05649-6. Commun Biol. 2023. PMID: 38082099 Free PMC article.
-
Deep mutational scanning: A versatile tool in systematically mapping genotypes to phenotypes.Front Genet. 2023 Jan 12;14:1087267. doi: 10.3389/fgene.2023.1087267. eCollection 2023. Front Genet. 2023. PMID: 36713072 Free PMC article. Review.
-
Mammalian Antigen Display for Pandemic Countermeasures.Methods Mol Biol. 2024;2762:191-216. doi: 10.1007/978-1-0716-3666-4_12. Methods Mol Biol. 2024. PMID: 38315367
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

