Micro-electron diffraction structure of the aggregation-driving N terminus of Drosophila neuronal protein Orb2A reveals amyloid-like β-sheets
- PMID: 35988647
- PMCID: PMC9556795
- DOI: 10.1016/j.jbc.2022.102396
Micro-electron diffraction structure of the aggregation-driving N terminus of Drosophila neuronal protein Orb2A reveals amyloid-like β-sheets
Abstract
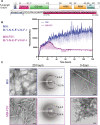
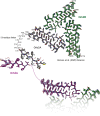
Amyloid protein aggregation is commonly associated with progressive neurodegenerative diseases, however not all amyloid fibrils are pathogenic. The neuronal cytoplasmic polyadenylation element binding protein is a regulator of synaptic mRNA translation and has been shown to form functional amyloid aggregates that stabilize long-term memory. In adult Drosophila neurons, the cytoplasmic polyadenylation element binding homolog Orb2 is expressed as 2 isoforms, of which the Orb2B isoform is far more abundant, but the rarer Orb2A isoform is required to initiate Orb2 aggregation. The N terminus is a distinctive feature of the Orb2A isoform and is critical for its aggregation. Intriguingly, replacement of phenylalanine in the fifth position of Orb2A with tyrosine (F5Y) in Drosophila impairs stabilization of long-term memory. The structure of endogenous Orb2B fibers was recently determined by cryo-EM, but the structure adopted by fibrillar Orb2A is less certain. Here we use micro-electron diffraction to determine the structure of the first 9 N-terminal residues of Orb2A, at a resolution of 1.05 Å. We find that this segment (which we term M9I) forms an amyloid-like array of parallel in-register β-sheets, which interact through side chain interdigitation of aromatic and hydrophobic residues. Our structure provides an explanation for the decreased aggregation observed for the F5Y mutant and offers a hypothesis for how the addition of a single atom (the tyrosyl oxygen) affects long-term memory. We also propose a structural model of Orb2A that integrates our structure of the M9I segment with the published Orb2B cryo-EM structure.
Keywords: amyloid; cytoplasmic polyadenylation element binding (CPEB) protein; electron microscopy; functional amyloid; intrinsically disordered protein; micro-electron diffraction (micro-ED); orb2; protein aggregation; protein structure.
Copyright © 2022. Published by Elsevier Inc.
Conflict of interest statement
Conflict of interest D.S.E. is a SAB member and equity holder in ADRx, Inc.
Figures


Similar articles
-
Identification and Structural Characterization of the N-terminal Amyloid Core of Orb2 isoform A.Sci Rep. 2016 Dec 6;6:38265. doi: 10.1038/srep38265. Sci Rep. 2016. PMID: 27922050 Free PMC article.
-
The Functional Amyloid Orb2A Binds to Lipid Membranes.Biophys J. 2017 Jul 11;113(1):37-47. doi: 10.1016/j.bpj.2017.05.039. Biophys J. 2017. PMID: 28700922 Free PMC article.
-
Calmodulin binds the N-terminus of the functional amyloid Orb2A inhibiting fibril formation.PLoS One. 2022 Jan 13;17(1):e0259872. doi: 10.1371/journal.pone.0259872. eCollection 2022. PLoS One. 2022. PMID: 35025866 Free PMC article.
-
Implications of the Orb2 Amyloid Structure in Huntington's Disease.Int J Mol Sci. 2020 Sep 21;21(18):6910. doi: 10.3390/ijms21186910. Int J Mol Sci. 2020. PMID: 32967102 Free PMC article. Review.
-
Cytoplasmic polyadenylation element binding proteins in development, health, and disease.Annu Rev Cell Dev Biol. 2014;30:393-415. doi: 10.1146/annurev-cellbio-101011-155831. Epub 2014 Jul 14. Annu Rev Cell Dev Biol. 2014. PMID: 25068488 Review.
Cited by
-
Recent Advances in Amyloids Structural Studies and Thin Film Applications.Molecules. 2025 Jul 9;30(14):2908. doi: 10.3390/molecules30142908. Molecules. 2025. PMID: 40733175 Free PMC article. Review.
-
Long-Term Memory Formation in Drosophila Depends on the 3'UTR of CPEB Gene orb2.Cells. 2023 Jan 14;12(2):318. doi: 10.3390/cells12020318. Cells. 2023. PMID: 36672258 Free PMC article.
-
Single-molecule visualization determines conformational substate ensembles in β-sheet-rich peptide fibrils.Sci Adv. 2023 Jul 7;9(27):eadg7943. doi: 10.1126/sciadv.adg7943. Epub 2023 Jul 5. Sci Adv. 2023. PMID: 37406110 Free PMC article.
-
The current role and evolution of X-ray crystallography in drug discovery and development.Expert Opin Drug Discov. 2023 Jul-Dec;18(11):1221-1230. doi: 10.1080/17460441.2023.2246881. Epub 2023 Aug 17. Expert Opin Drug Discov. 2023. PMID: 37592849 Free PMC article. Review.
-
Divergent evolution of low-complexity regions in the vertebrate CPEB protein family.Front Bioinform. 2025 Mar 20;5:1491735. doi: 10.3389/fbinf.2025.1491735. eCollection 2025. Front Bioinform. 2025. PMID: 40182702 Free PMC article.
References
-
- Eisenberg D.S., Sawaya M.R. Structural studies of amyloid proteins at the molecular level. Annu. Rev. Biochem. 2017;86:69–95. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

