Celastrol: A lead compound that inhibits SARS-CoV-2 replication, the activity of viral and human cysteine proteases, and virus-induced IL-6 secretion
- PMID: 35989498
- PMCID: PMC9539158
- DOI: 10.1002/ddr.21982
Celastrol: A lead compound that inhibits SARS-CoV-2 replication, the activity of viral and human cysteine proteases, and virus-induced IL-6 secretion
Abstract
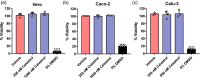
The global emergence of coronavirus disease 2019 (COVID-19) has caused substantial human casualties. Clinical manifestations of this disease vary from asymptomatic to lethal, and the symptomatic form can be associated with cytokine storm and hyperinflammation. In face of the urgent demand for effective drugs to treat COVID-19, we have searched for candidate compounds using in silico approach followed by experimental validation. Here we identified celastrol, a pentacyclic triterpene isolated from Tripterygium wilfordii Hook F, as one of the best compounds out of 39 drug candidates. Celastrol reverted the gene expression signature from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-infected cells and irreversibly inhibited the recombinant forms of the viral and human cysteine proteases involved in virus invasion, such as Mpro (main protease), PLpro (papain-like protease), and recombinant human cathepsin L. Celastrol suppressed SARS-CoV-2 replication in human and monkey cell lines and decreased interleukin-6 (IL-6) secretion in the SARS-CoV-2-infected human cell line. Celastrol acted in a concentration-dependent manner, with undetectable signs of cytotoxicity, and inhibited in vitro replication of the parental and SARS-CoV-2 variant. Therefore, celastrol is a promising lead compound to develop new drug candidates to face COVID-19 due to its ability to suppress SARS-CoV-2 replication and IL-6 production in infected cells.
Keywords: COVID-19; SARS-CoV-2; celastrol; cysteine protease inhibition; molecular docking.
© 2022 Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Olive-Derived Triterpenes Suppress SARS COV-2 Main Protease: A Promising Scaffold for Future Therapeutics.Molecules. 2021 May 1;26(9):2654. doi: 10.3390/molecules26092654. Molecules. 2021. PMID: 34062737 Free PMC article.
-
Interrelated Mechanism by Which the Methide Quinone Celastrol, Obtained from the Roots of Tripterygium wilfordii, Inhibits Main Protease 3CLpro of COVID-19 and Acts as Superoxide Radical Scavenger.Int J Mol Sci. 2020 Dec 4;21(23):9266. doi: 10.3390/ijms21239266. Int J Mol Sci. 2020. PMID: 33291769 Free PMC article.
-
Interaction of small molecules with the SARS-CoV-2 papain-like protease: In silico studies and in vitro validation of protease activity inhibition using an enzymatic inhibition assay.J Mol Graph Model. 2021 May;104:107851. doi: 10.1016/j.jmgm.2021.107851. Epub 2021 Jan 26. J Mol Graph Model. 2021. PMID: 33556646 Free PMC article.
-
SARS-CoV-2 Mpro: A Potential Target for Peptidomimetics and Small-Molecule Inhibitors.Biomolecules. 2021 Apr 19;11(4):607. doi: 10.3390/biom11040607. Biomolecules. 2021. PMID: 33921886 Free PMC article. Review.
-
Targeting SARS-CoV-2 viral proteases as a therapeutic strategy to treat COVID-19.J Med Virol. 2021 May;93(5):2722-2734. doi: 10.1002/jmv.26814. Epub 2021 Feb 9. J Med Virol. 2021. PMID: 33475167 Free PMC article. Review.
Cited by
-
Tripterin liposome relieves severe acute respiratory syndrome as a potent COVID-19 treatment.Signal Transduct Target Ther. 2022 Dec 24;7(1):399. doi: 10.1038/s41392-022-01283-6. Signal Transduct Target Ther. 2022. PMID: 36566328 Free PMC article.
-
Evaluation of Celastrol Antiviral Activity Against Equid Alphaherpesvirus Type 8 Infection.Viruses. 2025 Feb 28;17(3):347. doi: 10.3390/v17030347. Viruses. 2025. PMID: 40143276 Free PMC article.
-
Exploring the Antiviral Potential of Esters of Cinnamic Acids with Quercetin.Viruses. 2024 Apr 24;16(5):665. doi: 10.3390/v16050665. Viruses. 2024. PMID: 38793547 Free PMC article.
-
Lessons learnt from broad-spectrum coronavirus antiviral drug discovery.Expert Opin Drug Discov. 2024 Sep;19(9):1023-1041. doi: 10.1080/17460441.2024.2385598. Epub 2024 Jul 30. Expert Opin Drug Discov. 2024. PMID: 39078037 Free PMC article. Review.
-
Neuroprotective Agents with Therapeutic Potential for COVID-19.Biomolecules. 2023 Oct 27;13(11):1585. doi: 10.3390/biom13111585. Biomolecules. 2023. PMID: 38002267 Free PMC article. Review.
References
-
- Banerjee, A. , Nasir, J. A. , Budylowski, P. , Yip, L. , Aftanas, P. , Christie, N. , Ghalami, A. , Baid, K. , Raphenya, A. R. , Hirota, J. A. , Miller, M. S. , McGeer, A. J. , Ostrowski, M. , Kozak, R. A. , McArthur, A. G. , Mossman, K. , & Mubareka, S . (2020). Isolation, sequence, infectivity, and replication kinetics of severe acute respiratory syndrome coronavirus 2. Emerging Infectious Diseases, 26(9), 2054–2063. 10.3201/eid2609.201495 - DOI - PMC - PubMed
-
- Bassanini, I. , Parapini, S. , Ferrandi, E. E. , Gabriele, E. , Basilico, N. , Taramelli, D. , & Sparatore, A. (2021). Design, synthesis and in vitro investigation of novel basic celastrol carboxamides as bio‐inspired leishmanicidal agents endowed with inhibitory activity against leishmania Hsp90. Biomolecules, 11(1), 56. 10.3390/biom11010056 - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

