Single cell transcriptomics and TCR reconstruction reveal CD4 T cell response to MHC-II-restricted APOB epitope in human cardiovascular disease
- PMID: 35990517
- PMCID: PMC9383695
- DOI: 10.1038/s44161-022-00063-3
Single cell transcriptomics and TCR reconstruction reveal CD4 T cell response to MHC-II-restricted APOB epitope in human cardiovascular disease
Abstract
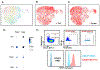
Atherosclerosis is accompanied by a CD4 T cell response to apolipoprotein B (APOB). Major Histocompatibility Complex (MHC)-II tetramers can be used to isolate antigen-specific CD4 T cells by flow sorting. Here, we produce, validate and use an MHC-II tetramer, DRB1*07:01 APOB-p18, to sort APOB-p18-specific cells from peripheral blood mononuclear cell samples from 8 DRB1*07:01+ women with and without subclinical cardiovascular disease (sCVD). Single cell RNA sequencing showed that transcriptomes of tetramer+ cells were between regulatory and memory T cells in healthy women and moved closer to memory T cells in women with sCVD. TCR sequencing of tetramer+ cells showed clonal expansion and V and J segment usage similar to those found in regulatory T cells. These findings suggest that APOB-specific regulatory T cells may switch to a more memory-like phenotype in women with atherosclerosis. Mouse studies showed that such switched cells promote atherosclerosis.
Conflict of interest statement
DISCLOSURES There are no conflicts of interest.
Figures



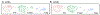




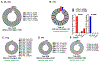





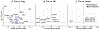
Similar articles
-
Regulatory CD4+ T Cells Recognize Major Histocompatibility Complex Class II Molecule-Restricted Peptide Epitopes of Apolipoprotein B.Circulation. 2018 Sep 11;138(11):1130-1143. doi: 10.1161/CIRCULATIONAHA.117.031420. Circulation. 2018. PMID: 29588316 Free PMC article.
-
Immunodominant MHC-II (Major Histocompatibility Complex II) Restricted Epitopes in Human Apolipoprotein B.Circ Res. 2022 Jul 22;131(3):258-276. doi: 10.1161/CIRCRESAHA.122.321116. Epub 2022 Jun 29. Circ Res. 2022. PMID: 35766025 Free PMC article.
-
Single-cell transcriptomes and T cell receptors of vaccine-expanded apolipoprotein B-specific T cells.Front Cardiovasc Med. 2023 Jan 5;9:1076808. doi: 10.3389/fcvm.2022.1076808. eCollection 2022. Front Cardiovasc Med. 2023. PMID: 36684560 Free PMC article.
-
[MHC tetramers: tracking specific immunity].Acta Med Croatica. 2003;57(4):255-9. Acta Med Croatica. 2003. PMID: 14639858 Review. Croatian.
-
Use of HLA class II tetramers in tracking antigen-specific T cells and mapping T-cell epitopes.Methods. 2003 Mar;29(3):282-8. doi: 10.1016/s1046-2023(02)00350-x. Methods. 2003. PMID: 12725793 Review.
Cited by
-
Hyperlipidaemia elicits an atypical, T helper 1-like CD4+ T-cell response: a key role for very low-density lipoprotein.Eur Heart J Open. 2023 Mar 3;3(2):oead013. doi: 10.1093/ehjopen/oead013. eCollection 2023 Mar. Eur Heart J Open. 2023. PMID: 36969380 Free PMC article.
-
Low-dose interleukin-2 induces clonal expansion of BACH2-repressed effector regulatory T cells following acute coronary syndrome.Nat Cardiovasc Res. 2025 Jun;4(6):727-739. doi: 10.1038/s44161-025-00652-y. Epub 2025 Jun 3. Nat Cardiovasc Res. 2025. PMID: 40461772 Free PMC article.
-
Apolipoprotein B-containing lipoproteins in atherogenesis.Nat Rev Cardiol. 2025 Jun;22(6):399-413. doi: 10.1038/s41569-024-01111-0. Epub 2025 Jan 2. Nat Rev Cardiol. 2025. PMID: 39743565 Review.
-
Loss of effector Treg signature in APOB-reactive CD4+ T cells in patients with coronary artery disease.Nat Cardiovasc Res. 2025 Jul;4(7):841-856. doi: 10.1038/s44161-025-00671-9. Epub 2025 Jun 18. Nat Cardiovasc Res. 2025. PMID: 40533521
-
Targeting immune cell recruitment in atherosclerosis.Nat Rev Cardiol. 2024 Nov;21(11):824-840. doi: 10.1038/s41569-024-01023-z. Epub 2024 Apr 25. Nat Rev Cardiol. 2024. PMID: 38664575 Review.
References
-
- Kimura T, Kobiyama K, Winkels H, Tse K, Miller J, Vassallo M, Wolf D, Ryden C, Orecchioni M, Dileepan T, Jenkins MK, James EA, Kwok WW, Hanna DB, Kaplan RC, Strickler HD, Durkin HG, Kassaye SG, Karim R, Tien PC, Landay AL, Gange SJ, Sidney J, Sette A, Ley K. Regulatory CD4+ T Cells Recognize Major Histocompatibility Complex Class II Molecule–Restricted Peptide Epitopes of Apolipoprotein B. Circulation. 2018;138:1130–1143. - PMC - PubMed
-
- Wolf D, Gerhardt T, Winkels H, Michel NA, Pramod AB, Ghosheh Y, Brunel S, Buscher K, Miller J, McArdle S, Baas L, Kobiyama K, Vassallo M, Ehinger E, Dileepan T, Ali A, Schell M, Mikulski Z, Sidler D, Kimura T, Sheng X, Horstmann H, Hansen S, Mitre LS, Stachon P, Hilgendorf I, Gaddis DE, Hedrick C, Benedict CA, Peters B, Zirlik A, Sette A, Ley K. Pathogenic Autoimmunity in Atherosclerosis Evolves From Initially Protective Apolipoprotein B100-Reactive CD4+ T-Regulatory Cells. Circulation. 2020;142:1279–1293. - PMC - PubMed
Grants and funding
- U01 HL146245/HL/NHLBI NIH HHS/United States
- P01 HL136275/HL/NHLBI NIH HHS/United States
- U01 HL146208/HL/NHLBI NIH HHS/United States
- R35 HL145241/HL/NHLBI NIH HHS/United States
- U01 HL146192/HL/NHLBI NIH HHS/United States
- U01 HL146242/HL/NHLBI NIH HHS/United States
- U01 HL146193/HL/NHLBI NIH HHS/United States
- R01 HL148094/HL/NHLBI NIH HHS/United States
- U01 HL146194/HL/NHLBI NIH HHS/United States
- U01 HL146241/HL/NHLBI NIH HHS/United States
- P30 AI027767/AI/NIAID NIH HHS/United States
- P30 AI050409/AI/NIAID NIH HHS/United States
- U01 HL146333/HL/NHLBI NIH HHS/United States
- U01 HL146205/HL/NHLBI NIH HHS/United States
- P30 MH116867/MH/NIMH NIH HHS/United States
- P30 AI073961/AI/NIAID NIH HHS/United States
- U01 HL146201/HL/NHLBI NIH HHS/United States
- U01 HL146204/HL/NHLBI NIH HHS/United States
- S10 OD021831/OD/NIH HHS/United States
- U01 HL146202/HL/NHLBI NIH HHS/United States
- UL1 TR001881/TR/NCATS NIH HHS/United States
- UL1 TR000004/TR/NCATS NIH HHS/United States
- U01 HL146240/HL/NHLBI NIH HHS/United States
- U01 HL146203/HL/NHLBI NIH HHS/United States
- K01 HL137557/HL/NHLBI NIH HHS/United States
- UL1 TR003098/TR/NCATS NIH HHS/United States
- P30 AI050410/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
