COVID-19 diagnosis using deep learning neural networks applied to CT images
- PMID: 35990616
- PMCID: PMC9389263
- DOI: 10.3389/frai.2022.919672
COVID-19 diagnosis using deep learning neural networks applied to CT images
Abstract
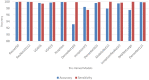
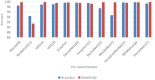
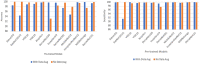
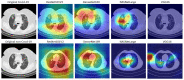
COVID-19, a deadly and highly contagious virus, caused the deaths of millions of individuals around the world. Early detection of the virus can reduce the virus transmission and fatality rate. Many deep learning (DL) based COVID-19 detection methods have been proposed, but most are trained on either small, incomplete, noisy, or imbalanced datasets. Many are also trained on a small number of COVID-19 samples. This study tackles these concerns by introducing DL-based solutions for COVID-19 diagnosis using computerized tomography (CT) images and 12 cutting-edge DL pre-trained models with acceptable Top-1 accuracy. All the models are trained on 9,000 COVID-19 samples and 5,000 normal images, which is higher than the COVID-19 images used in most studies. In addition, while most of the research used X-ray images for training, this study used CT images. CT scans capture blood arteries, bones, and soft tissues more effectively than X-Ray. The proposed techniques were evaluated, and the results show that NASNetLarge produced the best classification accuracy, followed by InceptionResNetV2 and DenseNet169. The three models achieved an accuracy of 99.86, 99.79, and 99.71%, respectively. Moreover, DenseNet121 and VGG16 achieved the best sensitivity, while InceptionV3 and InceptionResNetV2 achieved the best specificity. DenseNet121 and VGG16 attained a sensitivity of 99.94%, while InceptionV3 and InceptionResNetV2 achieved a specificity of 100%. The models are compared to those designed in three existing studies, and they produce better results. The results show that deep neural networks have the potential for computer-assisted COVID-19 diagnosis. We hope this study will be valuable in improving the decisions and accuracy of medical practitioners when diagnosing COVID-19. This study will assist future researchers in minimizing the repetition of analysis and identifying the ideal network for their tasks.
Keywords: COVID-19 diagnosis; CT images; convolutional neural network; deep learning networks; pre-trained models.
Copyright © 2022 Akinyelu and Blignaut.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
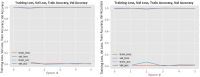
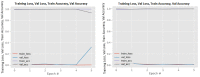
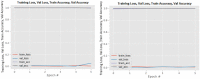
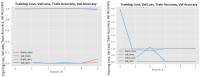
Figures




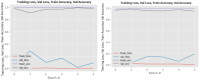
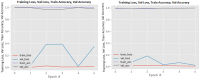

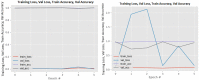
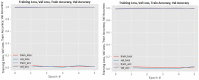


References
-
- An B. J., Xu P., Harmon S., Turkbey S. A., Sanford E. B., Amalou T. H., et al. (2020). CT Images in COVID-19 [Data set]. Available online at: https://wiki.cancerimagingarchive.net/display/Public/CT+Images+in+COVID-19 (accessed January 5, 2022).
-
- Ardakani A. A., Kanafi A. R., Acharya U. R., Khadem N., Mohammadi A. (2020). Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput. Biol. Med. 121, 103795. 10.1016/j.compbiomed.2020.103795 - DOI - PMC - PubMed
-
- Chollet F. (2017). “Xception: deep learning with depthwise separable convolutions,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 1251–1258.
-
- Faisal A., Dharma T. N. A., Ferani W., Widi P. G., Rizka S. F. (2021). Comparative study of VGG16 and MobileNetv2 for masked face recognition. J. Ilm. Tek. Elektro Komput. 7, 230–237. 10.26555/jiteki.v7i2.20758 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

