CRISPR screening identifies T cell-intrinsic regulators of CD3-bispecific antibody responses
- PMID: 35990699
- PMCID: PMC9388929
- DOI: 10.3389/fimmu.2022.909979
CRISPR screening identifies T cell-intrinsic regulators of CD3-bispecific antibody responses
Abstract
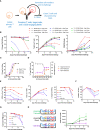
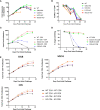
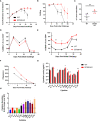
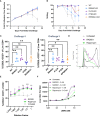
CD3-engaging bispecific antibodies (BsAbs) enable the formation of an immune synapse between T cells and tumor cells, resulting in robust target cell killing not dependent on a preexisting tumor specific T cell receptor. While recent studies have shed light on tumor cell-specific factors that modulate BsAb sensitivity, the T cell-intrinsic determinants of BsAb efficacy and response durability are poorly understood. To better clarify the genes that shape BsAb-induced T cell responses, we conducted targeted analyses and a large-scale unbiased in vitro CRISPR/Cas9-based screen to identify negative regulators of BsAb-induced T cell proliferation. These analyses revealed that CD8+ T cells are dependent on CD4+ T cell-derived signaling factors in order to achieve sustained killing in vitro. Moreover, the mammalian target of rapamycin (mTOR) pathway and several other candidate genes were identified as intrinsic regulators of BsAb-induced T cell proliferation and/or activation, highlighting promising approaches to enhancing the utility of these potent therapeutics.
Keywords: Bispecific antibodies; CRISPR screening; EP300; T cells; immunotherapy; mTOR.
Copyright © 2022 Molony, Funk, Trabucco, Corcoran, Ruddy, Varadarajan, Elliot, Piquet, Lam, Meyer, Wang, Kurtulus and Lu.
Conflict of interest statement
Authors RM, TF, GT, EC, DR, MP, JL, HW, and HL are currently employed by the Novartis Institutes for Biomedical Research (NIBR). The following authors were previously employed by NIBR when they were involved with the work described herein: MV (currently employed by Arbor Biotechnologies), GE (currently employed by Foghorn Therapeutics), MM (currently employed by Bristol Myers Squibb), and SK (currently employed by 2seventy Bio).
Figures

References
-
- Weiner GJ, Kostelny SA, Hillstrom JR, Cole MS, Link BK, Wang SL, et al. . The role of T cell activation in anti-CD3 x antitumor bispecific antibody therapy. J Immunol (1994) 152(5):2385–92. - PubMed
-
- Topp MS, Gökbuget N, Zugmaier G, Klappers P, Stelljes M, Neumann S, et al. . Phase II trial of the anti-CD19 bispecific T cell–engager blinatumomab shows hematologic and molecular remissions in patients with relapsed or refractory b-precursor acute lymphoblastic leukemia. J Clin Oncol (2014) 32(36):4134–40. doi: 10.1200/JCO.2014.56.3247 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

