Effects of the Nature of the Metal Ion, Protein and Substrate on the Catalytic Center in Matrix Metalloproteinase-1: Insights from Multilevel MD, QM/MM and QM Studies
- PMID: 35991515
- PMCID: PMC9387770
- DOI: 10.1002/cphc.202100680
Effects of the Nature of the Metal Ion, Protein and Substrate on the Catalytic Center in Matrix Metalloproteinase-1: Insights from Multilevel MD, QM/MM and QM Studies
Abstract
Matrix metalloproteinase-1 (MMP-1) is a Zn(II) dependent endopeptidase involved in the degradation of collagen, the most abundant structural protein in the extracellular matrix of connective tissues and the human body. Herein we performed a multilevel computational analysis including molecular dynamics (MD), combined quantum mechanics/molecular mechanics (QM/MM), and quantum mechanics (QM) calculations to characterize the structure and geometry of the catalytic Zn(II) within the MMP-1 protein environment in comparison to crystallographic and spectroscopic data. The substrate's removal fine-tuned impact on the conformational dynamics and geometry of the catalytic Zn(II) center was also explored. Finally, the study examined the effect of substituting catalytic Zn(II) by Co(II) on the overall structure and dynamics of the MMP-1 THP complex and specifically on the geometry of the catalytic metal center. Overall our QM/MM and QM studies were in good agreement with the MM description of the Zn(II) centers in the MD simulations.
Keywords: Matrix metalloproteinases; Molecular dynamics; QM/MM calculations; Zn(II) containing enzymes.
Conflict of interest statement
Conflict of Interest The authors declare no conflict of interest.
Figures

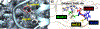

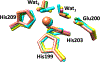

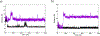
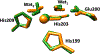
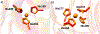
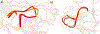
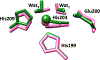
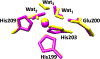

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

