Integrative Analysis Between Genome-Wide Association Study and Expression Quantitative Trait Loci Reveals Bovine Muscle Gene Expression Regulatory Polymorphisms Associated With Intramuscular Fat and Backfat Thickness
- PMID: 35991540
- PMCID: PMC9386181
- DOI: 10.3389/fgene.2022.935238
Integrative Analysis Between Genome-Wide Association Study and Expression Quantitative Trait Loci Reveals Bovine Muscle Gene Expression Regulatory Polymorphisms Associated With Intramuscular Fat and Backfat Thickness
Abstract
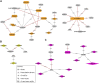
Understanding the architecture of gene expression is fundamental to unravel the molecular mechanisms regulating complex traits in bovine, such as intramuscular fat content (IMF) and backfat thickness (BFT). These traits are economically important for the beef industry since they affect carcass and meat quality. Our main goal was to identify gene expression regulatory polymorphisms within genomic regions (QTL) associated with IMF and BFT in Nellore cattle. For that, we used RNA-Seq data from 193 Nellore steers to perform SNP calling analysis. Then, we combined the RNA-Seq SNP and a high-density SNP panel to obtain a new dataset for further genome-wide association analysis (GWAS), totaling 534,928 SNPs. GWAS was performed using the Bayes B model. Twenty-one relevant QTL were associated with our target traits. The expression quantitative trait loci (eQTL) analysis was performed using Matrix eQTL with the complete SNP dataset and 12,991 genes, revealing a total of 71,033 cis and 36,497 trans-eQTL (FDR < 0.05). Intersecting with QTL for IMF, we found 231 eQTL regulating the expression levels of 117 genes. Within those eQTL, three predicted deleterious SNPs were identified. We also identified 109 eQTL associated with BFT and affecting the expression of 54 genes. This study revealed genomic regions and regulatory SNPs associated with fat deposition in Nellore cattle. We highlight the transcription factors FOXP4, FOXO3, ZSCAN2, and EBF4, involved in lipid metabolism-related pathways. These results helped us to improve our knowledge about the genetic architecture behind important traits in cattle.
Keywords: Nellore cattle; RNA-Seq; SNP; backfat thickness; carcass and meat quality; expression quantitative trait loci; intramuscular fat content.
Copyright © 2022 Silva-Vignato, Cesar, Afonso, Moreira, Poleti, Petrini, Garcia, Clemente, Mourão, Regitano and Coutinho.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- AOCS (2004). “Rapid Determination of Oil/fat Utilizing High Temperature Solvent Extraction,”. AOCS Official Procedure Am 5-04 in Official Methods and Recommended Practices of the AOCS (Champaign, IL: American Oil Chemists Society, AOCS; ).
-
- Benjamini Y., Hochberg Y. (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 57 (1), 289–300. 10.1111/j.2517-6161.1995.tb02031.x - DOI
-
- Borges B. O., Curi R. A., Baldi F., Feitosa F. L. B., Andrade W. B. F. d., Albuquerque L. G. d., et al. (2014). Polymorphisms in Candidate Genes and Their Association with Carcass Traits and Meat Quality in Nellore Cattle. Pesq. Agropec. Bras. 49 (5), 364–371. 10.1590/S0100-204X2014000500006 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

