Examining SNP-SNP interactions and risk of clinical outcomes in colorectal cancer using multifactor dimensionality reduction based methods
- PMID: 35991579
- PMCID: PMC9385108
- DOI: 10.3389/fgene.2022.902217
Examining SNP-SNP interactions and risk of clinical outcomes in colorectal cancer using multifactor dimensionality reduction based methods
Abstract
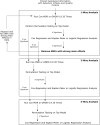
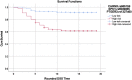
Background: SNP interactions may explain the variable outcome risk among colorectal cancer patients. Examining SNP interactions is challenging, especially with large datasets. Multifactor Dimensionality Reduction (MDR)-based programs may address this problem. Objectives: 1) To compare two MDR-based programs for their utility; and 2) to apply these programs to sets of MMP and VEGF-family gene SNPs in order to examine their interactions in relation to colorectal cancer survival outcomes. Methods: This study applied two data reduction methods, Cox-MDR and GMDR 0.9, to study one to three way SNP interactions. Both programs were run using a 5-fold cross validation step and the top models were verified by permutation testing. Prognostic associations of the SNP interactions were verified using multivariable regression methods. Eight datasets, including SNPs from MMP family genes (n = 201) and seven sets of VEGF-family interaction networks (n = 1,517 SNPs) were examined. Results: ∼90 million potential interactions were examined. Analyses in the MMP and VEGF gene family datasets found several novel 1- to 3-way SNP interactions. These interactions were able to distinguish between the patients with different outcome risks (regression p-values 0.03-2.2E-09). The strongest association was detected for a 3-way interaction including CHRM3.rs665159_EPN1.rs6509955_PTGER3.rs1327460 variants. Conclusion: Our work demonstrates the utility of data reduction methods while identifying potential prognostic markers in colorectal cancer.
Keywords: MDR; MMPs; SNP-SNP interactions; VEGF family; colorectal cancer; prognostic markers; protein interaction network.
Copyright © 2022 Curtis, Yu, Carey, Parfrey, Yilmaz and Savas.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Afzal S., Gusella M., Vainer B., Vogel U. B., Andersen J. T., Broedbaek K., et al. (2011b). Combinations of polymorphisms in genes involved in the 5-Fluorouracil metabolism pathway are associated with gastrointestinal toxicity in chemotherapy-treated colorectal cancer patients. Clin. Cancer Res. 17, 3822–3829. 10.1158/1078-0432.CCR-11-0304 - DOI - PubMed
-
- Archives Ensembl. Available at: http://useast.ensembl.org/info/website/archives/index.html (Accessed January 20, 2021).
LinkOut - more resources
Full Text Sources

