Diversity of clonal complex 22 methicillin-resistant Staphylococcus aureus isolates in Kuwait hospitals
- PMID: 35992657
- PMCID: PMC9386227
- DOI: 10.3389/fmicb.2022.970924
Diversity of clonal complex 22 methicillin-resistant Staphylococcus aureus isolates in Kuwait hospitals
Abstract
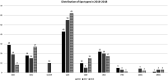
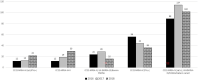
CC22-MRSA is a major MRSA lineage that is widely reported globally. To characterize CC22-MRSA for trends in antibiotic resistance and emergence of variants, a total of 636 CC22 isolates identified by DNA microarray in 2016 (n = 195), 2017 (n = 227) and 2018 (n = 214) were investigated further using staphylococcal protein A (spa) typing and multilocus sequence typing. The isolates belonged to 109 spa types dominated by t223 (n = 160), t032 (n = 60), t852 (n = 59), t005 (n = 56) and t309 (n = 30) and 10 sequence types (STs) dominated by ST22 (85.5%). Genotypes CC22-MRSA-IV [tst1+]; CC22-MRSA-IV UK-EMRSA-15/Barnim EMRSA variants, CC22-MRSA-IV [PVL+], CC22-MRSA-IV [tst1+/PVL+] and CC22-MRSA-IV + V constituted >50% of the isolates. An increase from 2016 to 2018 were shown in isolates belonging to spa types t223 (43 to 62), t032 (18 to 27) and t309 (10 to 15) and genotypes CC22-MRSA-IV [tst1+] (89 to 102), CC22-MRSA-IV + V (12 to 30) and CC22-MRSA-IV [tst1+/PVL+] (12 to 22). Ninety-nine CC22-MRSA isolates were multi-resistant to three or more antibiotic classes with 76.7% of them belonging to CC22-MRSA-IV [PVL+] and CC22-MRSA-IV [tst1+/PVL+]. The study revealed an ongoing domination of the CC22-MRSA-[tst1+] genotype and the emergence of new clones bearing SCCmec IV + V and multiply resistant variants.
Keywords: CC22-MRSA; DNA microarray; antibiotic resistance; genotypes; spa typing.
Copyright © 2022 Boswihi, Verghese and Udo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Al Laham N., Mediavilla J. R., Chen L., Abdelateef N., Elamreen F. A., Ginocchio C. C., et al. . (2015). MRSA clonal complex 22 strains harboring toxic shock syndrome toxin (TSST-1) are endemic in the primary hospital in Gaza, Palestine. PLoS One. 10:e0120008. doi: 10.1371/journal.pone.0120008, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases