Application of a novel lytic phage vB_EcoM_SQ17 for the biocontrol of Enterohemorrhagic Escherichia coli O157:H7 and Enterotoxigenic E. coli in food matrices
- PMID: 35992713
- PMCID: PMC9389114
- DOI: 10.3389/fmicb.2022.929005
Application of a novel lytic phage vB_EcoM_SQ17 for the biocontrol of Enterohemorrhagic Escherichia coli O157:H7 and Enterotoxigenic E. coli in food matrices
Abstract
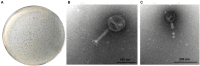
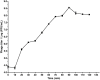
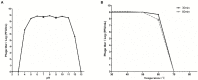
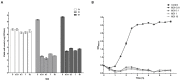
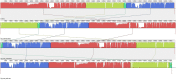
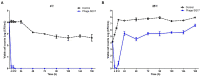
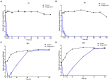
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 and Enterotoxigenic E. coli (ETEC) are important foodborne pathogens, causing serious food poisoning outbreaks worldwide. Bacteriophages, as novel antibacterial agents, have been increasingly exploited to control foodborne pathogens. In this study, a novel broad-host range lytic phage vB_EcoM_SQ17 (SQ17), was isolated, characterized, and evaluated for its potential to control bacterial counts in vitro and in three different food matrices (milk, raw beef, and fresh lettuce). Phage SQ17 was capable of infecting EHEC O157:H7, ETEC, and other E. coli strains. Morphology, one-step growth, and stability assay showed that phage SQ17 belongs to the Caudovirales order, Myoviridae family, and Mosigvirus genus. It has a short latent period of 10 min, a burst size of 71 PFU/infected cell, high stability between pH 4 to 12 as well as thermostability between 30°C and 60°C for 60 min. Genome sequencing analysis revealed that the genome of SQ17 does not contain any genes associated with antibiotic resistance, toxins, lysogeny, or virulence factors, indicating the potential safe application of phage SQ17 in the food industry. In Luria-Bertani (LB) medium, phage SQ17 significantly decreased the viable counts of EHEC O157:H7 by more than 2.40 log CFU/ml (p < 0.05) after 6 h of incubation at 37°C. Phage SQ17 showed great potential to be applied for biocontrol of EHEC O157:H7 in milk and raw beef. In fresh lettuce, treatment with SQ17 also resulted in significant reduction of viable cell counts of EHEC O157:H7 and ETEC at both 4°C and 25°C. Our results demonstrate that SQ17 is a good candidate for application as an EHEC O157:H7 and ETEC biocontrol agent in the processing stages of food production and food preservation.
Keywords: EHEC O157:H7; ETEC; biocontrol; foodborne pathogen; foods; phage.
Copyright © 2022 Zhou, Wan, Bao, Guo, Zhu, Zhang, Pang and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Efficacy of a lytic bacteriophage vB_EcoM_SQ17 against Enterohemorrhagic Escherichia coli O157:H7 and Enterotoxigenic E. coli biofilms on cucumber.Microb Pathog. 2024 Sep;194:106832. doi: 10.1016/j.micpath.2024.106832. Epub 2024 Jul 31. Microb Pathog. 2024. PMID: 39089511
-
Molecular characterization and safety properties of multi drug-resistant Escherichia coli O157:H7 bacteriophages.BMC Microbiol. 2024 Dec 19;24(1):528. doi: 10.1186/s12866-024-03691-w. BMC Microbiol. 2024. PMID: 39695941 Free PMC article.
-
CAM-21, a novel lytic phage with high specificity towards Escherichia coli O157:H7 in food products.Int J Food Microbiol. 2023 Feb 2;386:110026. doi: 10.1016/j.ijfoodmicro.2022.110026. Epub 2022 Nov 23. Int J Food Microbiol. 2023. PMID: 36444789
-
Animal health and foodborne pathogens: enterohaemorrhagic O157:H7 strains and other pathogenic Escherichia coli virotypes (EPEC, ETEC, EIEC, EHEC).Pol J Vet Sci. 2002;5(2):103-15. Pol J Vet Sci. 2002. PMID: 12189946 Review.
-
Safety Properties of Escherichia coli O157:H7 Specific Bacteriophages: Recent Advances for Food Safety.Foods. 2023 Oct 31;12(21):3989. doi: 10.3390/foods12213989. Foods. 2023. PMID: 37959107 Free PMC article. Review.
Cited by
-
Can Bacteriophages Be Effectively Utilized for Disinfection in Animal-Derived Food Products? A Systematic Review.Pathogens. 2025 Mar 16;14(3):291. doi: 10.3390/pathogens14030291. Pathogens. 2025. PMID: 40137775 Free PMC article.
-
Antibacterial effects and mechanisms of quercetin-β-cyclodextrin complex mediated photodynamic on Escherichia coli O157:H7.Arch Microbiol. 2024 Oct 23;206(11):445. doi: 10.1007/s00203-024-04175-1. Arch Microbiol. 2024. PMID: 39443369
-
Genome sequencing of Escherichia coli phage UFJF_EcSW4 reveals a novel lytic Kayfunavirus species.3 Biotech. 2025 Jan;15(1):10. doi: 10.1007/s13205-024-04172-7. Epub 2024 Dec 15. 3 Biotech. 2025. PMID: 39691801
-
A Review on Recent Trends in Bacteriophages for Post-Harvest Food Decontamination.Microorganisms. 2025 Feb 27;13(3):515. doi: 10.3390/microorganisms13030515. Microorganisms. 2025. PMID: 40142412 Free PMC article. Review.
-
Genomic analysis, culturing optimization, and characterization of Escherichia bacteriophage OSYSP, previously studied as effective pathogen control on fresh produce.Front Microbiol. 2024 Dec 9;15:1486333. doi: 10.3389/fmicb.2024.1486333. eCollection 2024. Front Microbiol. 2024. PMID: 39717272 Free PMC article.
References
-
- Bao H., Shahin K., Zhang Q., Zhang H., Wang Z., Zhou Y., et al. . (2019). Microbial pathogenesis morphologic and genomic characterization of a broad host range Salmonella enterica serovar Pullorum lytic phage vB_SPuM_SP116. Microbial Pthogenesis 136:103659. doi: 10.1016/j.micpath.2019.103659, PMID: - DOI - PubMed
-
- Carter C. D., Parks A., Abuladze T., Li M., Woolston J., Magnone J., et al. . (2012). Bacteriophage cocktail significantly reduces Escherichia coli O157:H7 contamination of lettuce and beef, but does not protect against recontamination. Bacteriophage 2, 178–185. doi: 10.4161/bact.22825, PMID: - DOI - PMC - PubMed
-
- Castro-rosas J., Cerna-cortés J. F., Méndez-reyes E., Lopez-hernandez D., Gómez-aldapa C. A., Estrada-garcia T. (2012). Presence of faecal coliforms, Escherichia coli and diarrheagenic E. coli pathotypes in ready-to-eat salads, from an area where crops are irrigated with untreated sewage water. Int. J. Food Microbiol. 156, 176–180. doi: 10.1016/j.ijfoodmicro.2012.03.025, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources

