Identification and characterization of N6-methyladenosine circular RNAs in the spinal cord of morphine-tolerant rats
- PMID: 35992914
- PMCID: PMC9388936
- DOI: 10.3389/fnins.2022.967768
Identification and characterization of N6-methyladenosine circular RNAs in the spinal cord of morphine-tolerant rats
Abstract
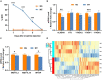
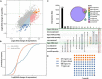
Morphine tolerance (MT) is a tricky problem, the mechanism of it is currently unknown. Circular RNAs (circRNAs) serve significant functions in the biological processes (BPs) of the central nervous system. N6-methyladenosine (m6A), as a key post-transcriptional modification of RNA, can regulate the metabolism and functions of circRNAs. Here we explore the patterns of m6A-methylation of circRNAs in the spinal cord of morphine-tolerant rats. In brief, we constructed a morphine-tolerant rat model, performed m6A epitranscriptomic microarray using RNA samples collected from the spinal cords of morphine-tolerant rats and normal saline rats, and implemented the bioinformatics analysis. In the spinal cord of morphine-tolerant rats, 120 circRNAs with different m6A modifications were identified, 54 of which were hypermethylated and 66 of which were hypomethylated. Functional analysis of these m6A circRNAs found some important pathways involved in the pathogenesis of MT, such as the calcium signaling pathway. In the m6A circRNA-miRNA networks, several critical miRNAs that participated in the occurrence and development of MT were discovered to bind to these m6A circRNAs, such as miR-873a-5p, miR-103-1-5p, miR-107-5p. M6A modification of circRNAs may be involved in the pathogenesis of MT. These findings may lead to new insights into the epigenetic etiology and pathology of MT.
Keywords: N6-methyladenosine; bioinformatics analysis; circRNAs; microarray; morphine tolerance.
Copyright © 2022 Xing, Deng, Shi, Dai, Ding, Song and Zou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Circular RNA expression profile in the spinal cord of morphine tolerated rats and screen of putative key circRNAs.Mol Brain. 2019 Sep 18;12(1):79. doi: 10.1186/s13041-019-0498-4. Mol Brain. 2019. PMID: 31533844 Free PMC article.
-
Genome-wide map of N6-methyladenosine circular RNAs identified in mice model of severe acute pancreatitis.World J Gastroenterol. 2021 Nov 21;27(43):7530-7545. doi: 10.3748/wjg.v27.i43.7530. World J Gastroenterol. 2021. PMID: 34887647 Free PMC article.
-
N6-methyladenosine methylation analysis of circRNAs in acquired middle ear cholesteatoma.Front Genet. 2024 Jun 24;15:1396720. doi: 10.3389/fgene.2024.1396720. eCollection 2024. Front Genet. 2024. PMID: 38978876 Free PMC article.
-
Advances in detecting N6-methyladenosine modification in circRNAs.Methods. 2022 Sep;205:234-246. doi: 10.1016/j.ymeth.2022.07.011. Epub 2022 Jul 22. Methods. 2022. PMID: 35878749 Review.
-
Novel insights into the interaction between N6-methyladenosine modification and circular RNA.Mol Ther Nucleic Acids. 2022 Jan 10;27:824-837. doi: 10.1016/j.omtn.2022.01.007. eCollection 2022 Mar 8. Mol Ther Nucleic Acids. 2022. PMID: 35141044 Free PMC article. Review.
Cited by
-
Circular RNA regulation and function in drug seeking phenotypes.Mol Cell Neurosci. 2023 Jun;125:103841. doi: 10.1016/j.mcn.2023.103841. Epub 2023 Mar 17. Mol Cell Neurosci. 2023. PMID: 36935046 Free PMC article. Review.
-
Orbitofrontal intronic circular RNA from Nrxn3 mediates reward learning and motivation for reward.Prog Neurobiol. 2024 Jan;232:102546. doi: 10.1016/j.pneurobio.2023.102546. Epub 2023 Nov 29. Prog Neurobiol. 2024. PMID: 38036039 Free PMC article.
-
Epigenetic Insights into Substance Use Disorder and Associated Psychiatric Conditions.Complex Psychiatry. 2025 Mar 3;11(1):12-36. doi: 10.1159/000544912. eCollection 2025 Jan-Dec. Complex Psychiatry. 2025. PMID: 40201238 Free PMC article. Review.
-
m6A modifications of circular RNAs in ischemia-induced retinal neovascularization.Int J Med Sci. 2023 Jan 22;20(2):254-261. doi: 10.7150/ijms.79409. eCollection 2023. Int J Med Sci. 2023. PMID: 36794165 Free PMC article.
-
tRNA epitranscriptomic alterations associated with opioid-induced reward-seeking and long-term opioid withdrawal in male mice.Neuropsychopharmacology. 2024 Jul;49(8):1276-1284. doi: 10.1038/s41386-024-01813-6. Epub 2024 Feb 8. Neuropsychopharmacology. 2024. PMID: 38332016 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

