The A to I editing landscape in melanoma and its relation to clinical outcome
- PMID: 35993275
- PMCID: PMC9415457
- DOI: 10.1080/15476286.2022.2110390
The A to I editing landscape in melanoma and its relation to clinical outcome
Abstract
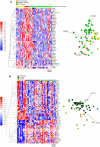
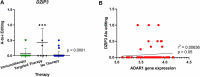
RNA editing refers to non-transient RNA modifications that occur after transcription and prior to translation by the ribosomes. RNA editing is more widespread in cancer cells than in non-transformed cells and is associated with tumorigenesis of various cancer tissues. However, RNA editing can also generate neo-antigens that expose tumour cells to host immunosurveillance. Global RNA editing in melanoma and its relevance to clinical outcome currently remain poorly characterized. The present study compared RNA editing as well as gene expression in tumour cell lines from melanoma patients of short or long metastasis-free survival, patients relapsing or not after immuno- and targeted therapy and tumours harbouring BRAF or NRAS mutations. Overall, our results showed that NTRK gene expression can be a marker of resistance to BRAF and MEK inhibition and gives some insights of candidate genes as potential biomarkers. In addition, this study revealed an increase in Adenosine-to-Inosine editing in Alu regions and in non-repetitive regions, including the hyperediting of the MOK and DZIP3 genes in relapsed tumour samples during targeted therapy and of the ZBTB11 gene in NRAS mutated melanoma cells. Therefore, RNA editing could be a promising tool for identifying predictive markers, tumour neoantigens and targetable pathways that could help in preventing relapses during immuno- or targeted therapies.
Keywords: A-to-I editing; ADAR; Alu sequences; Editing; immune checkpoint inhibitors; immunotherapy; melanoma.
Conflict of interest statement
There are no financial and non-financial competing interests
Figures


Similar articles
-
Evaluation of the Interplay between the ADAR Editome and Immunotherapy in Melanoma.Noncoding RNA. 2021 Jan 12;7(1):5. doi: 10.3390/ncrna7010005. Noncoding RNA. 2021. PMID: 33445472 Free PMC article.
-
MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: a non-randomised, open-label phase 2 study.Lancet Oncol. 2013 Mar;14(3):249-56. doi: 10.1016/S1470-2045(13)70024-X. Epub 2013 Feb 13. Lancet Oncol. 2013. PMID: 23414587 Clinical Trial.
-
MEK inhibition may increase survival of NRAS-mutated melanoma patients treated with checkpoint blockade: Results of a retrospective multicentre analysis of 364 patients.Eur J Cancer. 2018 Jul;98:10-16. doi: 10.1016/j.ejca.2018.04.010. Epub 2018 May 26. Eur J Cancer. 2018. PMID: 29843107
-
Targeted therapies and immune checkpoint inhibitors in the treatment of metastatic melanoma patients: a guide and update for pathologists.Pathology. 2016 Feb;48(2):194-202. doi: 10.1016/j.pathol.2015.12.010. Epub 2016 Jan 20. Pathology. 2016. PMID: 27020392 Review.
-
Targeted therapies in melanoma beyond BRAF: targeting NRAS-mutated and KIT-mutated melanoma.Curr Opin Oncol. 2020 Mar;32(2):79-84. doi: 10.1097/CCO.0000000000000606. Curr Opin Oncol. 2020. PMID: 31833955 Review.
Cited by
-
The diverse landscape of RNA modifications in cancer development and progression.Genes Genomics. 2025 Feb;47(2):135-155. doi: 10.1007/s13258-024-01601-y. Epub 2024 Dec 6. Genes Genomics. 2025. PMID: 39643826 Review.
-
RNA therapeutics.RNA Biol. 2024 Jan;21(1):1-2. doi: 10.1080/15476286.2022.2161704. Epub 2023 Jan 11. RNA Biol. 2024. PMID: 36629444 Free PMC article. No abstract available.
-
The role of ADAR1 through and beyond its editing activity in cancer.Cell Commun Signal. 2024 Jan 17;22(1):42. doi: 10.1186/s12964-023-01465-x. Cell Commun Signal. 2024. PMID: 38233935 Free PMC article. Review.
-
Whole tumour cell-based vaccines: tuning the instruments to orchestrate an optimal antitumour immune response.Br J Cancer. 2023 Sep;129(4):572-585. doi: 10.1038/s41416-023-02327-6. Epub 2023 Jun 24. Br J Cancer. 2023. PMID: 37355722 Free PMC article. Review.
-
RNA modifications and their role in gene expression.Front Mol Biosci. 2025 Apr 25;12:1537861. doi: 10.3389/fmolb.2025.1537861. eCollection 2025. Front Mol Biosci. 2025. PMID: 40351534 Free PMC article. Review.
References
-
- Paz-Yaacov N, Bazak L, Buchumenski I, et al. Elevated RNA editing activity is a major contributor to transcriptomic diversity in tumors. Cell Rep. 2015;13(2):267–276. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
