The Diversity and Functional Capacity of Microbes Associated with Coastal Macrophytes
- PMID: 35993708
- PMCID: PMC9601103
- DOI: 10.1128/msystems.00592-22
The Diversity and Functional Capacity of Microbes Associated with Coastal Macrophytes
Abstract
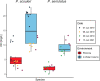
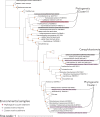
Coastal marine macrophytes exhibit some of the highest rates of primary productivity in the world. They have been found to host a diverse set of microbes, many of which may impact the biology of their hosts through metabolisms that are unique to microbial taxa. Here, we characterized the metabolic functions of macrophyte-associated microbial communities using metagenomes collected from 2 species of kelp (Laminaria setchellii and Nereocystis luetkeana) and 3 marine angiosperms (Phyllospadix scouleri, P. serrulatus, and Zostera marina), including the rhizomes of two surfgrass species (Phyllospadix spp.), the seagrass Zostera marina, and the sediments surrounding P. scouleri and Z. marina. Using metagenomic sequencing, we describe 63 metagenome-assembled genomes (MAGs) that potentially benefit from being associated with macrophytes and may contribute to macrophyte fitness through their metabolic activity. Host-associated metagenomes contained genes for the use of dissolved organic matter from hosts and vitamin (B1, B2, B7, B12) biosynthesis in addition to a range of nitrogen and sulfur metabolisms that recycle dissolved inorganic nutrients into forms more available to the host. The rhizosphere of surfgrass and seagrass contained genes for anaerobic microbial metabolisms, including nifH genes associated with nitrogen fixation, despite residing in a well-mixed and oxygenated environment. The range of oxygen environments engineered by macrophytes likely explains the diversity of both oxidizing and reducing microbial metabolisms and contributes to the functional capabilities of microbes and their influences on carbon and nitrogen cycling in nearshore ecosystems. IMPORTANCE Kelps, seagrasses, and surfgrasses are ecosystem engineers on rocky shorelines, where they show remarkably high levels of primary production. Through analysis of their associated microbial communities, we found a variety of microbial metabolisms that may benefit the host, including nitrogen metabolisms, sulfur oxidation, and the production of B vitamins. In turn, these microbes have the genetic capabilities to assimilate the dissolved organic compounds released by their macrophyte hosts. We describe a range of oxygen environments associated with surfgrass, including low-oxygen microhabitats in their rhizomes that host genes for nitrogen fixation. The tremendous productivity of coastal seaweeds and seagrasses is likely due in part to the activities of associated microbes, and an increased understanding of these associations is needed.
Keywords: host-microbiome relationships; kelp; macrophytes; marine microbiology; oxygen; seagrass; surfgrass.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Ammonification by kelp associated microbes increases ammonium availability.PLoS One. 2024 Mar 29;19(3):e0296622. doi: 10.1371/journal.pone.0296622. eCollection 2024. PLoS One. 2024. PMID: 38551914 Free PMC article.
-
Functional Insights into the Kelp Microbiome from Metagenome-Assembled Genomes.mSystems. 2022 Jun 28;7(3):e0142221. doi: 10.1128/msystems.01422-21. Epub 2022 Jun 1. mSystems. 2022. PMID: 35642511 Free PMC article.
-
Rhizosphere microbiomes are closely linked to seagrass species: a comparative study of three coastal seagrasses.Appl Environ Microbiol. 2024 Dec 18;90(12):e0175424. doi: 10.1128/aem.01754-24. Epub 2024 Nov 6. Appl Environ Microbiol. 2024. PMID: 39503478 Free PMC article.
-
The Seagrass Holobiont and Its Microbiome.Microorganisms. 2017 Dec 15;5(4):81. doi: 10.3390/microorganisms5040081. Microorganisms. 2017. PMID: 29244764 Free PMC article. Review.
-
Early anaerobic metabolisms.Philos Trans R Soc Lond B Biol Sci. 2006 Oct 29;361(1474):1819-34; discussion 1835-6. doi: 10.1098/rstb.2006.1906. Philos Trans R Soc Lond B Biol Sci. 2006. PMID: 17008221 Free PMC article. Review.
Cited by
-
Bacteria on the foundational kelp in kelp forest ecosystems: Insights from culturing, whole genome sequencing and metabolic assays.Environ Microbiol Rep. 2024 Jun;16(3):e13270. doi: 10.1111/1758-2229.13270. Environ Microbiol Rep. 2024. PMID: 38778582 Free PMC article.
-
Contrasting diversity and temporal patterns in leaf and root microbiome of two nearby temperate Zostera marina meadows.Environ Microbiome. 2025 Aug 5;20(1):98. doi: 10.1186/s40793-025-00760-z. Environ Microbiome. 2025. PMID: 40765019 Free PMC article.
-
Ammonification by kelp associated microbes increases ammonium availability.PLoS One. 2024 Mar 29;19(3):e0296622. doi: 10.1371/journal.pone.0296622. eCollection 2024. PLoS One. 2024. PMID: 38551914 Free PMC article.
-
Microbial associates of an endemic Mediterranean seagrass enhance the access of the host and the surrounding seawater to inorganic nitrogen under ocean acidification.Sci Rep. 2023 Nov 15;13(1):19996. doi: 10.1038/s41598-023-47126-4. Sci Rep. 2023. PMID: 37968499 Free PMC article.
-
Plants against cancer: the immune-boosting herbal microbiome: not of the plant, but in the plant. Basic concepts, introduction, and future resource for vaccine adjuvant discovery.Front Oncol. 2023 Jul 31;13:1180084. doi: 10.3389/fonc.2023.1180084. eCollection 2023. Front Oncol. 2023. PMID: 37588095 Free PMC article. Review.
References
-
- McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA 110:3229–3236. doi:10.1073/pnas.1218525110. - DOI - PMC - PubMed
-
- Duarte CM, Middelburg JJ, Caraco N. 2005. Major role of marine vegetation on the oceanic carbon cycle. Biogeosciences 2:1–8. doi:10.5194/bg-2-1-2005. - DOI
-
- Mohr W, Lehnen N, Ahmerkamp S, Marchant HK, Graf JS, Tschitschko B, Yilmaz P, Littmann S, Gruber-Vodicka H, Leisch N, Weber M, Lott C, Schubert CJ, Milucka J, Kuypers MMM. 2021. Terrestrial-type nitrogen-fixing symbiosis between seagrass and a marine bacterium. Nature 600:105–109. doi:10.1038/s41586-021-04063-4. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Miscellaneous

