Activation of Adaptive and Innate Immune Cells via Localized IL2 Cytokine Factories Eradicates Mesothelioma Tumors
- PMID: 35993913
- PMCID: PMC9713361
- DOI: 10.1158/1078-0432.CCR-22-1493
Activation of Adaptive and Innate Immune Cells via Localized IL2 Cytokine Factories Eradicates Mesothelioma Tumors
Abstract
Purpose: IL2 immunotherapy has the potential to elicit immune-mediated tumor lysis via activation of effector immune cells, but clinical utility is limited due to pharmacokinetic challenges as well as vascular leak syndrome and other life-threatening toxicities experienced by patients. We developed a safe and clinically translatable localized IL2 delivery system to boost the potency of therapy while minimizing systemic cytokine exposure.
Experimental design: We evaluated the therapeutic efficacy of IL2 cytokine factories in a mouse model of malignant mesothelioma. Changes in immune populations were analyzed using time-of-flight mass cytometry (CyTOF), and the safety and translatability of the platform were evaluated using complete blood counts and serum chemistry analysis.
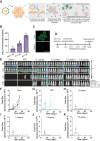
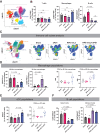
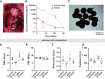
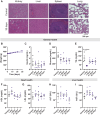
Results: IL2 cytokine factories enabled 150× higher IL2 concentrations in the local compartment with limited leakage into the systemic circulation. AB1 tumor burden was reduced by 80% after 1 week of monotherapy treatment, and 7 of 7 of animals exhibited tumor eradication without recurrence when IL2 cytokine factories were combined with anti-programmed cell death protein 1 (aPD1). Furthermore, CyTOF analysis showed an increase in CD69+CD44+ and CD69-CD44+CD62L- T cells, reduction of CD86-PD-L1- M2-like macrophages, and a corresponding increase in CD86+PD-L1+ M1-like macrophages and MHC-II+ dendritic cells after treatment. Finally, blood chemistry ranges in rodents demonstrated the safety of cytokine factory treatment and reinforced its potential for clinical use.
Conclusions: IL2 cytokine factories led to the eradication of aggressive mouse malignant mesothelioma tumors and protection from tumor recurrence, and increased the therapeutic efficacy of aPD1 checkpoint therapy. This study provides support for the clinical evaluation of this IL2-based delivery system. See related commentary by Palanki et al., p. 5010.
©2022 The Authors; Published by the American Association for Cancer Research.
Figures


Comment in
-
A (Controlled) Spill of IL2 for Localized Treatment of Mesothelioma.Clin Cancer Res. 2022 Dec 1;28(23):5010-5012. doi: 10.1158/1078-0432.CCR-22-2626. Clin Cancer Res. 2022. PMID: 36190329 Free PMC article.
References
-
- Nelson DB, Rice DC, Niu J, Atay S, Vaporciyan AA, Antonoff M, et al. Long-term survival outcomes of cancer-directed surgery for malignant pleural mesothelioma: propensity score matching analysis. J Clin Oncol 2017;35:3354–62. - PubMed
-
- Musk AW, Olsen N, Alfonso H, Reid A, Mina R, Franklin P, et al. Predicting survival in malignant mesothelioma. Eur Respir J 2011;38:1420–4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

