Exploiting breakdown in nonhost effector-target interactions to boost host disease resistance
- PMID: 35994659
- PMCID: PMC9436328
- DOI: 10.1073/pnas.2114064119
Exploiting breakdown in nonhost effector-target interactions to boost host disease resistance
Abstract
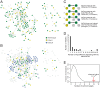
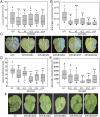
Plants are resistant to most microbial species due to nonhost resistance (NHR), providing broad-spectrum and durable immunity. However, the molecular components contributing to NHR are poorly characterised. We address the question of whether failure of pathogen effectors to manipulate nonhost plants plays a critical role in NHR. RxLR (Arg-any amino acid-Leu-Arg) effectors from two oomycete pathogens, Phytophthora infestans and Hyaloperonospora arabidopsidis, enhanced pathogen infection when expressed in host plants (Nicotiana benthamiana and Arabidopsis, respectively) but the same effectors performed poorly in distantly related nonhost pathosystems. Putative target proteins in the host plant potato were identified for 64 P. infestans RxLR effectors using yeast 2-hybrid (Y2H) screens. Candidate orthologues of these target proteins in the distantly related non-host plant Arabidopsis were identified and screened using matrix Y2H for interaction with RxLR effectors from both P. infestans and H. arabidopsidis. Few P. infestans effector-target protein interactions were conserved from potato to candidate Arabidopsis target orthologues (cAtOrths). However, there was an enrichment of H. arabidopsidis RxLR effectors interacting with cAtOrths. We expressed the cAtOrth AtPUB33, which unlike its potato orthologue did not interact with P. infestans effector PiSFI3, in potato and Nicotiana benthamiana. Expression of AtPUB33 significantly reduced P. infestans colonization in both host plants. Our results provide evidence that failure of pathogen effectors to interact with and/or correctly manipulate target proteins in distantly related non-host plants contributes to NHR. Moreover, exploiting this breakdown in effector-nonhost target interaction, transferring effector target orthologues from non-host to host plants is a strategy to reduce disease.
Keywords: effector-triggered susceptibility; host range; oomycete; plant immunity; plant–microbe interactions.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
-
- Jones J. D. G., Dangl J. L., The plant immune system. Nature 444, 323–329 (2006). - PubMed
-
- Win J., et al. , Effector biology of plant-associated organisms: Concepts and perspectives. Cold Spring Harb. Symp. Quant. Biol. 77, 235–247 (2012). - PubMed
-
- Panstruga R., Moscou M. J., What is the molecular basis of nonhost resistance? Mol. Plant Microbe Interact. 33, 1253–1264 (2020). - PubMed
-
- Uma B., Rani T. S., Podile A. R., Warriors at the gate that never sleep: Non-host resistance in plants. J. Plant Physiol. 168, 2141–2152 (2011). - PubMed
-
- Schulze-Lefert P., Panstruga R., A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci. 16, 117–125 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/G015244/1/UKRI | Biotechnology and Biological Sciences Research Council (BBSRC)
- BB/K018612/2/UKRI | Biotechnology and Biological Sciences Research Council (BBSRC)
- BB/N009967/1/UKRI | Biotechnology and Biological Sciences Research Council (BBSRC)
- BB/P020569/1/UKRI | Biotechnology and Biological Sciences Research Council (BBSRC)
- BB/S015663/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

