BAP1 maintains HIF-dependent interferon beta induction to suppress tumor growth in clear cell renal cell carcinoma
- PMID: 35995140
- PMCID: PMC9553033
- DOI: 10.1016/j.canlet.2022.215885
BAP1 maintains HIF-dependent interferon beta induction to suppress tumor growth in clear cell renal cell carcinoma
Abstract
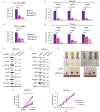
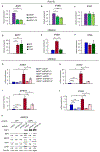
BRCA1-associated protein 1 (BAP1) is a deubiquitinase that is mutated in 10-15% of clear cell renal cell carcinomas (ccRCC). Despite the association between BAP1 loss and poor clinical outcome, the critical tumor suppressor function(s) of BAP1 in ccRCC remains unclear. Previously, we found that hypoxia-inducible factor 2α (HIF2α) and BAP1 activate interferon-stimulated gene factor 3 (ISGF3), a transcription factor activated by type I interferons and a tumor suppressor in ccRCC xenograft models. Here, we aimed to determine the mechanism(s) through which HIF and BAP1 regulate ISGF3. We found that in ccRCC cells, loss of the von Hippel-Lindau tumor suppressor (VHL) activated interferon beta (IFN-β) expression in a HIF2α-dependent manner. IFN-β was required for ISGF3 activation and suppressed the growth of Ren-02 tumors in xenografts. BAP1 enhanced the expression of IFN-β and stimulator of interferon genes (STING), both of which activate ISGF3. Both ISGF3 overexpression and STING agonist treatment increased ISGF3 activity and suppressed BAP1-deficient tumor growth in Ren-02 xenografts. Our results indicate that BAP1 loss reduces type I interferon signaling, and reactivating this pathway may be a novel therapeutic strategy for treating ccRCC.
Keywords: BAP1; HIF; Interferon; STING; ccRCC.
Copyright © 2022 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare no competing interests.
Figures






References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA: A Cancer Journal for Clinicians. 2021;71(1):7–33. - PubMed
-
- Kaelin WG. HIF2 Inhibitor Joins the Kidney Cancer Armamentarium. J Clin Oncol. 2018;36(9):908–10. - PubMed
-
- Choueiri TK, Kaelin WG. Targeting the HIF2-VEGF axis in renal cell carcinoma. Nature Medicine. 2020;26(10):1519–30. - PubMed
-
- Choueiri TK, Motzer RJ. Systemic Therapy for Metastatic Renal-Cell Carcinoma. New England Journal of Medicine. 2017;376(4):354–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

