The antileukemic activity of decitabine upon PML/RARA-negative AML blasts is supported by all-trans retinoic acid: in vitro and in vivo evidence for cooperation
- PMID: 35995769
- PMCID: PMC9395383
- DOI: 10.1038/s41408-022-00715-4
The antileukemic activity of decitabine upon PML/RARA-negative AML blasts is supported by all-trans retinoic acid: in vitro and in vivo evidence for cooperation
Abstract
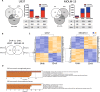
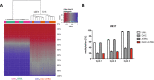
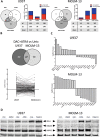
The prognosis of AML patients with adverse genetics, such as a complex, monosomal karyotype and TP53 lesions, is still dismal even with standard chemotherapy. DNA-hypomethylating agent monotherapy induces an encouraging response rate in these patients. When combined with decitabine (DAC), all-trans retinoic acid (ATRA) resulted in an improved response rate and longer overall survival in a randomized phase II trial (DECIDER; NCT00867672). The molecular mechanisms governing this in vivo synergism are unclear. We now demonstrate cooperative antileukemic effects of DAC and ATRA on AML cell lines U937 and MOLM-13. By RNA-sequencing, derepression of >1200 commonly regulated transcripts following the dual treatment was observed. Overall chromatin accessibility (interrogated by ATAC-seq) and, in particular, at motifs of retinoic acid response elements were affected by both single-agent DAC and ATRA, and enhanced by the dual treatment. Cooperativity regarding transcriptional induction and chromatin remodeling was demonstrated by interrogating the HIC1, CYP26A1, GBP4, and LYZ genes, in vivo gene derepression by expression studies on peripheral blood blasts from AML patients receiving DAC + ATRA. The two drugs also cooperated in derepression of transposable elements, more effectively in U937 (mutated TP53) than MOLM-13 (intact TP53), resulting in a "viral mimicry" response. In conclusion, we demonstrate that in vitro and in vivo, the antileukemic and gene-derepressive epigenetic activity of DAC is enhanced by ATRA.
© 2022. The Author(s).
Conflict of interest statement
HB received honoraria from BMS, Pierre Fabre Pharma, Servier, MSD and Novartis. ML is on the advisory boards of AbbVie, Astex Pharmaceuticals, Janssen, and Syros. He received research funding from Janssen and research support (study drug) from Cheplapharm and Janssen. The remaining authors declare no conflicts of interest.
Figures





References
-
- Klobuch S, Steinberg T, Bruni E, Mirbeth C, Heilmeier B, Ghibelli L, et al. Biomodulatory treatment with azacitidine, all-trans retinoic acid and pioglitazone induces differentiation of primary AML blasts into neutrophil like cells capable of ROS production and phagocytosis. Front Pharm. 2018;9:1380. doi: 10.3389/fphar.2018.01380. - DOI - PMC - PubMed
-
- Soriano AO, Yang H, Faderl S, Estrov Z, Giles F, Ravandi F, et al. Safety and clinical activity of the combination of 5-azacytidine, valproic acid, and all-trans retinoic acid in acute myeloid leukemia and myelodysplastic syndrome. Blood. 2007;110:2302–8. doi: 10.1182/blood-2007-03-078576. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

