Evolution of longitudinal division in multicellular bacteria of the Neisseriaceae family
- PMID: 35995772
- PMCID: PMC9395523
- DOI: 10.1038/s41467-022-32260-w
Evolution of longitudinal division in multicellular bacteria of the Neisseriaceae family
Abstract
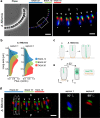
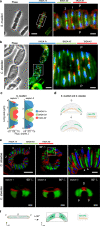
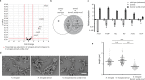
Rod-shaped bacteria typically elongate and divide by transverse fission. However, several bacterial species can form rod-shaped cells that divide longitudinally. Here, we study the evolution of cell shape and division mode within the family Neisseriaceae, which includes Gram-negative coccoid and rod-shaped species. In particular, bacteria of the genera Alysiella, Simonsiella and Conchiformibius, which can be found in the oral cavity of mammals, are multicellular and divide longitudinally. We use comparative genomics and ultrastructural microscopy to infer that longitudinal division within Neisseriaceae evolved from a rod-shaped ancestor. In multicellular longitudinally-dividing species, neighbouring cells within multicellular filaments are attached by their lateral peptidoglycan. In these bacteria, peptidoglycan insertion does not appear concentric, i.e. from the cell periphery to its centre, but as a medial sheet guillotining each cell. Finally, we identify genes and alleles associated with multicellularity and longitudinal division, including the acquisition of amidase-encoding gene amiC2, and amino acid changes in proteins including MreB and FtsA. Introduction of amiC2 and allelic substitution of mreB in a rod-shaped species that divides by transverse fission results in shorter cells with longer septa. Our work sheds light on the evolution of multicellularity and longitudinal division in bacteria, and suggests that members of the Neisseriaceae family may be good models to study these processes due to their morphological plasticity and genetic tractability.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

