The whole blood transcriptional regulation landscape in 465 COVID-19 infected samples from Japan COVID-19 Task Force
- PMID: 35995775
- PMCID: PMC9395416
- DOI: 10.1038/s41467-022-32276-2
The whole blood transcriptional regulation landscape in 465 COVID-19 infected samples from Japan COVID-19 Task Force
Abstract
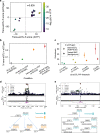
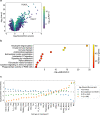
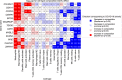
Coronavirus disease 2019 (COVID-19) is a recently-emerged infectious disease that has caused millions of deaths, where comprehensive understanding of disease mechanisms is still unestablished. In particular, studies of gene expression dynamics and regulation landscape in COVID-19 infected individuals are limited. Here, we report on a thorough analysis of whole blood RNA-seq data from 465 genotyped samples from the Japan COVID-19 Task Force, including 359 severe and 106 non-severe COVID-19 cases. We discover 1169 putative causal expression quantitative trait loci (eQTLs) including 34 possible colocalizations with biobank fine-mapping results of hematopoietic traits in a Japanese population, 1549 putative causal splice QTLs (sQTLs; e.g. two independent sQTLs at TOR1AIP1), as well as biologically interpretable trans-eQTL examples (e.g., REST and STING1), all fine-mapped at single variant resolution. We perform differential gene expression analysis to elucidate 198 genes with increased expression in severe COVID-19 cases and enriched for innate immune-related functions. Finally, we evaluate the limited but non-zero effect of COVID-19 phenotype on eQTL discovery, and highlight the presence of COVID-19 severity-interaction eQTLs (ieQTLs; e.g., CLEC4C and MYBL2). Our study provides a comprehensive catalog of whole blood regulatory variants in Japanese, as well as a reference for transcriptional landscapes in response to COVID-19 infection.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

