Spatially restricted drivers and transitional cell populations cooperate with the microenvironment in untreated and chemo-resistant pancreatic cancer
- PMID: 35995947
- PMCID: PMC9470535
- DOI: 10.1038/s41588-022-01157-1
Spatially restricted drivers and transitional cell populations cooperate with the microenvironment in untreated and chemo-resistant pancreatic cancer
Abstract
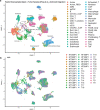
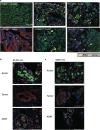
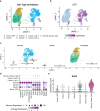
Pancreatic ductal adenocarcinoma is a lethal disease with limited treatment options and poor survival. We studied 83 spatial samples from 31 patients (11 treatment-naïve and 20 treated) using single-cell/nucleus RNA sequencing, bulk-proteogenomics, spatial transcriptomics and cellular imaging. Subpopulations of tumor cells exhibited signatures of proliferation, KRAS signaling, cell stress and epithelial-to-mesenchymal transition. Mapping mutations and copy number events distinguished tumor populations from normal and transitional cells, including acinar-to-ductal metaplasia and pancreatic intraepithelial neoplasia. Pathology-assisted deconvolution of spatial transcriptomic data identified tumor and transitional subpopulations with distinct histological features. We showed coordinated expression of TIGIT in exhausted and regulatory T cells and Nectin in tumor cells. Chemo-resistant samples contain a threefold enrichment of inflammatory cancer-associated fibroblasts that upregulate metallothioneins. Our study reveals a deeper understanding of the intricate substructure of pancreatic ductal adenocarcinoma tumors that could help improve therapy for patients with this disease.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures













References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J. Clin. 2022;72:7–33. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

