Brainprints: identifying individuals from magnetoencephalograms
- PMID: 35995976
- PMCID: PMC9395342
- DOI: 10.1038/s42003-022-03727-9
Brainprints: identifying individuals from magnetoencephalograms
Abstract
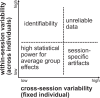
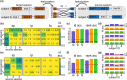
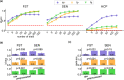
Magnetoencephalography (MEG) is used to study a wide variety of cognitive processes. Increasingly, researchers are adopting principles of open science and releasing their MEG data. While essential for reproducibility, sharing MEG data has unforeseen privacy risks. Individual differences may make a participant identifiable from their anonymized recordings. However, our ability to identify individuals based on these individual differences has not yet been assessed. Here, we propose interpretable MEG features to characterize individual difference. We term these features brainprints (brain fingerprints). We show through several datasets that brainprints accurately identify individuals across days, tasks, and even between MEG and Electroencephalography (EEG). Furthermore, we identify consistent brainprint components that are important for identification. We study the dependence of identifiability on the amount of data available. We also relate identifiability to the level of preprocessing and the experimental task. Our findings reveal specific aspects of individual variability in MEG. They also raise concerns about unregulated sharing of brain data, even if anonymized.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



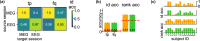

References
-
- Gorgolewski, K., Esteban, O., Schaefer, G., Wandell, B. & Poldrack, R. Openneuro-a Free Online Platform for Sharing and Analysis of Neuroimaging Data 1677 (Organization for Human Brain Mapping, 2017).
-
- Sweeney L. Simple demographics often identify people uniquely. Health (San Francisco) 2000;671:1–34.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

