BMP4 and Gremlin 1 regulate hepatic cell senescence during clinical progression of NAFLD/NASH
- PMID: 35995996
- PMCID: PMC9398907
- DOI: 10.1038/s42255-022-00620-x
BMP4 and Gremlin 1 regulate hepatic cell senescence during clinical progression of NAFLD/NASH
Abstract
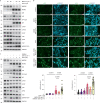
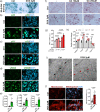
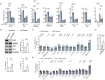
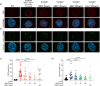
The role of hepatic cell senescence in human non-alcoholic fatty liver disease (NAFLD) and non-alcoholic steatohepatitis (NASH) is not well understood. To examine this, we performed liver biopsies and extensive characterization of 58 individuals with or without NAFLD/NASH. Here, we show that hepatic cell senescence is strongly related to NAFLD/NASH severity, and machine learning analysis identified senescence markers, the BMP4 inhibitor Gremlin 1 in liver and visceral fat, and the amount of visceral adipose tissue as strong predictors. Studies in liver cell spheroids made from human stellate and hepatocyte cells show BMP4 to be anti-senescent, anti-steatotic, anti-inflammatory and anti-fibrotic, whereas Gremlin 1, which is particularly highly expressed in visceral fat in humans, is pro-senescent and antagonistic to BMP4. Both senescence and anti-senescence factors target the YAP/TAZ pathway, making this a likely regulator of senescence and its effects. We conclude that senescence is an important driver of human NAFLD/NASH and that BMP4 and Gremlin 1 are novel therapeutic targets.
© 2022. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures










References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

