Reconsidering the czcD (NiCo) Riboswitch as an Iron Riboswitch
- PMID: 35996475
- PMCID: PMC9389577
- DOI: 10.1021/acsbiomedchemau.1c00069
Reconsidering the czcD (NiCo) Riboswitch as an Iron Riboswitch
Abstract
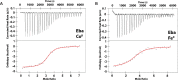
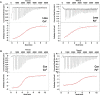
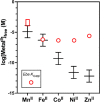
Recent work has proposed a new mechanism of bacterial iron regulation: riboswitches that undergo a conformational change in response to FeII. The czcD (NiCo) riboswitch was initially proposed to be specific for NiII and CoII, but we recently showed via a czcD-based fluorescent sensor that FeII is also a plausible physiological ligand for this riboswitch class. Here, we provide direct evidence that this riboswitch class responds to FeII. Isothermal titration calorimetry studies of the native czcD riboswitches from three organisms show no response to MnII, a weak response to ZnII, and similar dissociation constants (∼1 μM) and conformational responses for FeII, CoII, and NiII. Only the iron response is in the physiological concentration regime; the riboswitches' responses to CoII, NiII, and ZnII require 103-, 105-, and 106-fold higher "free" metal ion concentrations, respectively, than the typical availability of those metal ions in cells. By contrast, the "Sensei" RNA, recently claimed to be an iron-specific riboswitch, exhibits no response to FeII. Our results demonstrate that iron responsiveness is a conserved property of czcD riboswitches and clarify that this is the only family of iron-responsive riboswitch identified to date, setting the stage for characterization of their physiological function.
© 2022 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Iron-responsive riboswitches.Curr Opin Chem Biol. 2022 Jun;68:102135. doi: 10.1016/j.cbpa.2022.102135. Epub 2022 Apr 12. Curr Opin Chem Biol. 2022. PMID: 35427920 Free PMC article. Review.
-
The czcD (NiCo) Riboswitch Responds to Iron(II).Biochemistry. 2020 Apr 21;59(15):1508-1516. doi: 10.1021/acs.biochem.0c00074. Epub 2020 Apr 13. Biochemistry. 2020. PMID: 32250107
-
Structure-based investigations of the NAD+-II riboswitch.Nucleic Acids Res. 2023 Jan 11;51(1):54-67. doi: 10.1093/nar/gkac1227. Nucleic Acids Res. 2023. PMID: 36610789 Free PMC article.
-
A riboswitch-controlled TerC family transporter Alx tunes intracellular manganese concentration in Escherichia coli at alkaline pH.J Bacteriol. 2024 Jul 25;206(7):e0016824. doi: 10.1128/jb.00168-24. Epub 2024 Jun 13. J Bacteriol. 2024. PMID: 38869303 Free PMC article.
-
Riboswitch Mechanisms: New Tricks for an Old Dog.Biochemistry (Mosc). 2021 Aug;86(8):962-975. doi: 10.1134/S0006297921080071. Biochemistry (Mosc). 2021. PMID: 34488573 Free PMC article. Review.
Cited by
-
Metalation calculators for E. coli strain JM109 (DE3): aerobic, anaerobic, and hydrogen peroxide exposed cells cultured in LB media.Metallomics. 2022 Sep 1;14(9):mfac058. doi: 10.1093/mtomcs/mfac058. Metallomics. 2022. PMID: 35933161 Free PMC article.
-
Iron-responsive riboswitches.Curr Opin Chem Biol. 2022 Jun;68:102135. doi: 10.1016/j.cbpa.2022.102135. Epub 2022 Apr 12. Curr Opin Chem Biol. 2022. PMID: 35427920 Free PMC article. Review.
-
The current riboswitch landscape in Clostridioides difficile.Microbiology (Reading). 2024 Oct;170(10):001508. doi: 10.1099/mic.0.001508. Microbiology (Reading). 2024. PMID: 39405103 Free PMC article. Review.
-
Bacterial Metallostasis: Metal Sensing, Metalloproteome Remodeling, and Metal Trafficking.Chem Rev. 2024 Dec 25;124(24):13574-13659. doi: 10.1021/acs.chemrev.4c00264. Epub 2024 Dec 10. Chem Rev. 2024. PMID: 39658019 Free PMC article. Review.
-
Whole-Cell Biosensor for Iron Monitoring as a Potential Tool for Safeguarding Biodiversity in Polar Marine Environments.Mar Drugs. 2024 Jun 28;22(7):299. doi: 10.3390/md22070299. Mar Drugs. 2024. PMID: 39057408 Free PMC article. Review.
References
-
- Guth-Metzler R.; Bray M. S; Frenkel-Pinter M.; Suttapitugsakul S.; Montllor-Albalate C.; Bowman J. C; Wu R.; Reddi A. R; Okafor C D.; Glass J. B; Williams L. D. Cutting in-line with iron: ribosomal function and non-oxidative RNA cleavage. Nucleic Acids Res. 2020, 48, 8663–8674. 10.1093/nar/gkaa586. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

