Mutated KLF4(K409Q) in meningioma binds STRs and activates FGF3 gene expression
- PMID: 35996584
- PMCID: PMC9391581
- DOI: 10.1016/j.isci.2022.104839
Mutated KLF4(K409Q) in meningioma binds STRs and activates FGF3 gene expression
Abstract
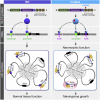
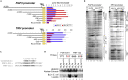
Krüppel-like factor 4 (KLF4) is a transcription factor that has been proven necessary for both induction and maintenance of pluripotency and self-renewal. Whole-genome sequencing defined a unique mutation in KLF4 (KLF4K409Q) in human meningiomas. However, the molecular mechanism of this tumor-specific KLF4 mutation is unknown. Using genome-wide high-throughput and focused quantitative transcriptional approaches in human cell lines, primary meningeal cells, and meningioma tumor tissue, we found that a change in the evolutionarily conserved DNA-binding domain of KLF4 alters its DNA recognition preference, resulting in a shift in downstream transcriptional activity. In the KLF4K409Q-specific targets, the normally silent fibroblast growth factor 3 (FGF3) is activated. We demonstrated a neomorphic function of KLF4K409Q in stimulating FGF3 transcription through binding to its promoter and in using short tandem repeats (STRs) located within the locus as enhancers.
Keywords: Cancer; Molecular biology; Transcriptomics.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Al-Mahdawi S., Pinto R.M., Ismail O., Varshney D., Lymperi S., Sandi C., Trabzuni D., Pook M. The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum. Mol. Genet. 2008;17:735–746. - PubMed
-
- Andrews S., Hamarneh G., Saad A. Fast random walker with priors using precomputation for interactive medical image segmentation. Med. Image Comput. Comput. Assist. Interv. 2010;13:9–16. - PubMed
-
- Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

