Integrative omics approaches for biosynthetic pathway discovery in plants
- PMID: 35997060
- PMCID: PMC9491492
- DOI: 10.1039/d2np00032f
Integrative omics approaches for biosynthetic pathway discovery in plants
Abstract
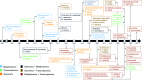
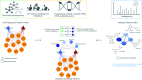
Covering: up to 2022With the emergence of large amounts of omics data, computational approaches for the identification of plant natural product biosynthetic pathways and their genetic regulation have become increasingly important. While genomes provide clues regarding functional associations between genes based on gene clustering, metabolome mining provides a foundational technology to chart natural product structural diversity in plants, and transcriptomics has been successfully used to identify new members of their biosynthetic pathways based on coexpression. Thus far, most approaches utilizing transcriptomics and metabolomics have been targeted towards specific pathways and use one type of omics data at a time. Recent technological advances now provide new opportunities for integration of multiple omics types and untargeted pathway discovery. Here, we review advances in plant biosynthetic pathway discovery using genomics, transcriptomics, and metabolomics, as well as recent efforts towards omics integration. We highlight how transcriptomics and metabolomics provide complementary information to link genes to metabolites, by associating temporal and spatial gene expression levels with metabolite abundance levels across samples, and by matching mass-spectral features to enzyme families. Furthermore, we suggest that elucidation of gene regulatory networks using time-series data may prove useful for efforts to unwire the complexities of biosynthetic pathway components based on regulatory interactions and events.
Conflict of interest statement
J. J. J. v. d. H. is a member of the Scientific Advisory Board of NAICONS Srl., Milano, Italy. M. H. M. is a member of the Scientific Advisory Board of Hexagon Bio and co-founder of Design Pharmaceuticals.
Figures







Similar articles
-
Toward the Heterologous Biosynthesis of Plant Natural Products: Gene Discovery and Characterization.ACS Synth Biol. 2021 Nov 19;10(11):2784-2795. doi: 10.1021/acssynbio.1c00315. Epub 2021 Nov 10. ACS Synth Biol. 2021. PMID: 34757715 Review.
-
Pairing omics to decode the diversity of plant specialized metabolism.Curr Opin Plant Biol. 2024 Dec;82:102657. doi: 10.1016/j.pbi.2024.102657. Epub 2024 Nov 10. Curr Opin Plant Biol. 2024. PMID: 39527852 Review.
-
Unlocking plant bioactive pathways: omics data harnessing and machine learning assisting.Curr Opin Biotechnol. 2024 Jun;87:103135. doi: 10.1016/j.copbio.2024.103135. Epub 2024 May 9. Curr Opin Biotechnol. 2024. PMID: 38728826 Review.
-
Integrative metabolo-genomics suggests a biosynthetic pathway for tetrangulol in Streptomyces sp. KL110A.World J Microbiol Biotechnol. 2025 Mar 11;41(3):101. doi: 10.1007/s11274-025-04298-7. World J Microbiol Biotechnol. 2025. PMID: 40064729 Free PMC article.
-
Enhanced correlation-based linking of biosynthetic gene clusters to their metabolic products through chemical class matching.Microbiome. 2023 Jan 23;11(1):13. doi: 10.1186/s40168-022-01444-3. Microbiome. 2023. PMID: 36691088 Free PMC article.
Cited by
-
Inventa: A computational tool to discover structural novelty in natural extracts libraries.Front Mol Biosci. 2022 Nov 11;9:1028334. doi: 10.3389/fmolb.2022.1028334. eCollection 2022. Front Mol Biosci. 2022. PMID: 36438653 Free PMC article.
-
The role of statistics in advancing nitric oxide research in plant biology: from data analysis to mechanistic insights.Front Plant Sci. 2025 Jul 1;16:1597030. doi: 10.3389/fpls.2025.1597030. eCollection 2025. Front Plant Sci. 2025. PMID: 40666296 Free PMC article. Review.
-
Chinese olive (Canarium album Rauesch.): a critical review on its nutritional value, phytochemical composition, health benefits, and practical applications.Front Pharmacol. 2023 Nov 29;14:1275113. doi: 10.3389/fphar.2023.1275113. eCollection 2023. Front Pharmacol. 2023. PMID: 38094884 Free PMC article. Review.
-
A Review of Transcriptomics and Metabolomics in Plant Quality and Environmental Response: From Bibliometric Analysis to Science Mapping and Future Trends.Metabolites. 2024 May 8;14(5):272. doi: 10.3390/metabo14050272. Metabolites. 2024. PMID: 38786749 Free PMC article. Review.
-
Discovering Dynamic Plant Enzyme Complexes in Yeast for Kratom Alkaloid Pathway Identification.Angew Chem Int Ed Engl. 2023 Sep 18;62(38):e202307995. doi: 10.1002/anie.202307995. Epub 2023 Aug 15. Angew Chem Int Ed Engl. 2023. PMID: 37549372 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

