X-Ray Scattering Reveals Two Mechanisms of Cellulose Microfibril Degradation by Filamentous Fungi
- PMID: 35997493
- PMCID: PMC9469724
- DOI: 10.1128/aem.00995-22
X-Ray Scattering Reveals Two Mechanisms of Cellulose Microfibril Degradation by Filamentous Fungi
Abstract
Mushroom-forming fungi (Agaricomycetes) employ enzymatic and nonenzymatic cellulose degradation mechanisms, the latter presumably relying on Fenton-generated radicals. The effects of the two mechanisms on the cellulose microfibrils structure remain poorly understood. We examined cellulose degradation caused by litter decomposers and wood decomposers, including brown-rot and white-rot fungi and one fungus with uncertain wood decay type, by combining small- and wide-angle X-ray scattering. We also examined the effects of commercial enzymes and Fenton-generated radicals on cellulose using the same method. We detected two main degradation or modification mechanisms. The first characterized the mechanism used by most fungi and resembled enzymatic cellulose degradation, causing simultaneous microfibril thinning and decreased crystalline cellulose. The second mechanism was detected in one brown-rot fungus and one litter decomposer and was characterized by patchy amorphogenesis of crystalline cellulose without substantial thinning of the fibers. This pattern did not resemble the effect of Fenton-generated radicals, suggesting a more complex mechanism is involved in the destruction of cellulose crystallinity by fungi. Furthermore, our results showed a mismatch between decay classifications and cellulose degradation patterns and that even within litter decomposers two degradation mechanisms were found, suggesting higher functional diversity under current ecological classifications of fungi. IMPORTANCE Cellulose degradation by fungi plays a fundamental role in terrestrial carbon cycling, but the mechanisms by which fungi cope with the crystallinity of cellulose are not fully understood. We used X-ray scattering to analyze how fungi, a commercial enzyme mix, and a Fenton reaction-generated radical alter the crystalline structure of cellulose. Our data revealed two mechanisms involved in crystalline cellulose degradation by fungi: one that results in the thinning of the cellulose fibers, resembling the enzymatic degradation of cellulose, and one that involves amorphogenesis of crystalline cellulose by yet-unknown pathways, resulting in a patchy-like degradation pattern. These results pave the way to a deeper understanding of cellulose degradation and the development of novel ways to utilize crystalline cellulose.
Keywords: Fenton chemistry; X-ray scattering; biodegradation; brown-rot fungus; cellulose; filamentous fungi; litter decomposer; white-rot fungus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

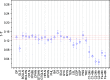
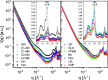

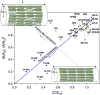
Similar articles
-
Uncovering the hidden diversity of litter-decomposition mechanisms in mushroom-forming fungi.ISME J. 2020 Aug;14(8):2046-2059. doi: 10.1038/s41396-020-0667-6. Epub 2020 May 7. ISME J. 2020. PMID: 32382073 Free PMC article.
-
Lignocellulosic polysaccharides and lignin degradation by wood decay fungi: the relevance of nonenzymatic Fenton-based reactions.J Ind Microbiol Biotechnol. 2011 Apr;38(4):541-55. doi: 10.1007/s10295-010-0798-2. Epub 2010 Aug 14. J Ind Microbiol Biotechnol. 2011. PMID: 20711629
-
Differences in crystalline cellulose modification due to degradation by brown and white rot fungi.Fungal Biol. 2012 Oct;116(10):1052-63. doi: 10.1016/j.funbio.2012.07.009. Epub 2012 Aug 8. Fungal Biol. 2012. PMID: 23063184
-
Lignin-modifying enzymes in filamentous basidiomycetes--ecological, functional and phylogenetic review.J Basic Microbiol. 2010 Feb;50(1):5-20. doi: 10.1002/jobm.200900338. J Basic Microbiol. 2010. PMID: 20175122 Review.
-
Biodegradation of lignocellulosics: microbial, chemical, and enzymatic aspects of the fungal attack of lignin.Int Microbiol. 2005 Sep;8(3):195-204. Int Microbiol. 2005. PMID: 16200498 Review.
Cited by
-
X-ray scattering based scanning tomography for imaging and structural characterization of cellulose in plants.J Synchrotron Radiat. 2024 Jul 1;31(Pt 4):936-947. doi: 10.1107/S1600577524004387. Epub 2024 Jun 25. J Synchrotron Radiat. 2024. PMID: 38917018 Free PMC article.
-
Impact of Agroforestry Practices on Soil Microbial Diversity and Nutrient Cycling in Atlantic Rainforest Cocoa Systems.Int J Mol Sci. 2024 Oct 22;25(21):11345. doi: 10.3390/ijms252111345. Int J Mol Sci. 2024. PMID: 39518901 Free PMC article.
References
-
- Chen H. 2014. Biotechnology of lignocellulose: theory and practice, p 25–71. Springer, Dordrecht, Netherlands.
-
- Grigoriev IV, Cullen D, Goodwin SB, Hibbett D, Jeffries TW, Kubicek CP, Kuske C, Magnuson JK, Martin F, Spatafora JW, Tsang A, Baker SE. 2011. Fueling the future with fungal genomics. Mycology 2:192–209.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

