Genome assembly of the chemosynthetic endosymbiont of the hydrothermal vent snail Alviniconcha adamantis from the Mariana Arc
- PMID: 35997584
- PMCID: PMC9526052
- DOI: 10.1093/g3journal/jkac220
Genome assembly of the chemosynthetic endosymbiont of the hydrothermal vent snail Alviniconcha adamantis from the Mariana Arc
Abstract
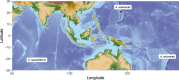
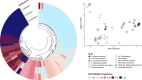
Chemosynthetic animal-microbe symbioses sustain hydrothermal vent communities in the global deep sea. In the Indo-Pacific Ocean, hydrothermal ecosystems are often dominated by gastropod species of the genus Alviniconcha, which live in association with chemosynthetic Gammaproteobacteria or Campylobacteria. While the symbiont genomes of most extant Alviniconcha species have been sequenced, no genome information is currently available for the gammaproteobacterial endosymbiont of Alviniconcha adamantis-a comparatively shallow living species that is thought to be the ancestor to all other present Alviniconcha lineages. Here, we report the first genome sequence for the symbiont of A. adamantis from the Chamorro Seamount at the Mariana Arc. Our phylogenomic analyses show that the A. adamantis symbiont is most closely related to Chromatiaceae endosymbionts of the hydrothermal vent snails Alviniconcha strummeri and Chrysomallon squamiferum, but represents a distinct bacterial species or possibly genus. Overall, the functional capacity of the A. adamantis symbiont appeared to be similar to other chemosynthetic Gammaproteobacteria, though several flagella and chemotaxis genes were detected, which are absent in other gammaproteobacterial Alviniconcha symbionts. These differences might suggest potential contrasts in symbiont transmission dynamics, host recognition, or nutrient transfer. Furthermore, an abundance of genes for ammonia transport and urea usage could indicate adaptations to the oligotrophic waters of the Mariana region, possibly via recycling of host- and environment-derived nitrogenous waste products. This genome assembly adds to the growing genomic resources for chemosynthetic bacteria from hydrothermal vents and will be valuable for future comparative genomic analyses assessing gene content evolution in relation to environment and symbiotic lifestyles.
Keywords: Alviniconcha adamantis; Mariana Arc; chemosynthetic symbiosis; hydrothermal vents.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America.
Figures

Similar articles
-
Allopatric and Sympatric Drivers of Speciation in Alviniconcha Hydrothermal Vent Snails.Mol Biol Evol. 2020 Dec 16;37(12):3469-3484. doi: 10.1093/molbev/msaa177. Mol Biol Evol. 2020. PMID: 32658967 Free PMC article.
-
Endosymbionts of Metazoans Dwelling in the PACManus Hydrothermal Vent: Diversity and Potential Adaptive Features Revealed by Genome Analysis.Appl Environ Microbiol. 2020 Oct 15;86(21):e00815-20. doi: 10.1128/AEM.00815-20. Print 2020 Oct 15. Appl Environ Microbiol. 2020. PMID: 32859597 Free PMC article.
-
Physiological dynamics of chemosynthetic symbionts in hydrothermal vent snails.ISME J. 2020 Oct;14(10):2568-2579. doi: 10.1038/s41396-020-0707-2. Epub 2020 Jul 2. ISME J. 2020. PMID: 32616905 Free PMC article.
-
Life in the Dark: Phylogenetic and Physiological Diversity of Chemosynthetic Symbioses.Annu Rev Microbiol. 2021 Oct 8;75:695-718. doi: 10.1146/annurev-micro-051021-123130. Epub 2021 Aug 5. Annu Rev Microbiol. 2021. PMID: 34351792 Review.
-
The microbiomes of deep-sea hydrothermal vents: distributed globally, shaped locally.Nat Rev Microbiol. 2019 May;17(5):271-283. doi: 10.1038/s41579-019-0160-2. Nat Rev Microbiol. 2019. PMID: 30867583 Review.
References
-
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W.. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27(4):578–579. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
