Expression and subcellular localization of USH1C/harmonin in human retina provides insights into pathomechanisms and therapy
- PMID: 35997788
- PMCID: PMC9851744
- DOI: 10.1093/hmg/ddac211
Expression and subcellular localization of USH1C/harmonin in human retina provides insights into pathomechanisms and therapy
Abstract
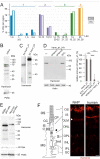
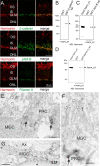
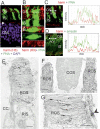
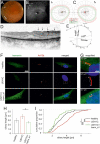
Usher syndrome (USH) is the most common form of hereditary deaf-blindness in humans. USH is a complex genetic disorder, assigned to three clinical subtypes differing in onset, course and severity, with USH1 being the most severe. Rodent USH1 models do not reflect the ocular phenotype observed in human patients to date; hence, little is known about the pathophysiology of USH1 in the human eye. One of the USH1 genes, USH1C, exhibits extensive alternative splicing and encodes numerous harmonin protein isoforms that function as scaffolds for organizing the USH interactome. RNA-seq analysis of human retinae uncovered harmonin_a1 as the most abundant transcript of USH1C. Bulk RNA-seq analysis and immunoblotting showed abundant expression of harmonin in Müller glia cells (MGCs) and retinal neurons. Furthermore, harmonin was localized in the terminal endfeet and apical microvilli of MGCs, presynaptic region (pedicle) of cones and outer segments (OS) of rods as well as at adhesive junctions between MGCs and photoreceptor cells (PRCs) in the outer limiting membrane (OLM). Our data provide evidence for the interaction of harmonin with OLM molecules in PRCs and MGCs and rhodopsin in PRCs. Subcellular expression and colocalization of harmonin correlate with the clinical phenotype observed in USH1C patients. We also demonstrate that primary cilia defects in USH1C patient-derived fibroblasts could be reverted by the delivery of harmonin_a1 transcript isoform. Our studies thus provide novel insights into PRC cell biology, USH1C pathophysiology and development of gene therapy treatment(s).
© The Author(s) 2022. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures



Similar articles
-
Identification of Unexpected Pathomechanisms Underlying the Human Usher Syndrome.Adv Exp Med Biol. 2025;1468:171-175. doi: 10.1007/978-3-031-76550-6_28. Adv Exp Med Biol. 2025. PMID: 39930191 Review.
-
Scaffold protein harmonin (USH1C) provides molecular links between Usher syndrome type 1 and type 2.Hum Mol Genet. 2005 Dec 15;14(24):3933-43. doi: 10.1093/hmg/ddi417. Epub 2005 Nov 21. Hum Mol Genet. 2005. PMID: 16301216
-
Molecular basis of human Usher syndrome: deciphering the meshes of the Usher protein network provides insights into the pathomechanisms of the Usher disease.Exp Eye Res. 2006 Jul;83(1):97-119. doi: 10.1016/j.exer.2005.11.010. Epub 2006 Mar 20. Exp Eye Res. 2006. PMID: 16545802 Review.
-
Photoreceptor expression of the Usher syndrome type 1 protein protocadherin 15 (USH1F) and its interaction with the scaffold protein harmonin (USH1C).Mol Vis. 2005 May 12;11:347-55. Mol Vis. 2005. PMID: 15928608
-
Early disruption of photoreceptor cell architecture and loss of vision in a humanized pig model of usher syndromes.EMBO Mol Med. 2022 Apr 7;14(4):e14817. doi: 10.15252/emmm.202114817. Epub 2022 Mar 7. EMBO Mol Med. 2022. PMID: 35254721 Free PMC article.
Cited by
-
Pathogenic Variants in USH1G/SANS Alter Protein Interaction with Pre-RNA Processing Factors PRPF6 and PRPF31 of the Spliceosome.Int J Mol Sci. 2023 Dec 18;24(24):17608. doi: 10.3390/ijms242417608. Int J Mol Sci. 2023. PMID: 38139438 Free PMC article.
-
Single-molecule fluorescence microscopy reveals regulatory mechanisms of MYO7A-driven cargo transport in stereocilia of live inner ear hair cells.Nat Commun. 2025 Sep 1;16(1):8149. doi: 10.1038/s41467-025-63102-0. Nat Commun. 2025. PMID: 40890108 Free PMC article.
-
Identification of Unexpected Pathomechanisms Underlying the Human Usher Syndrome.Adv Exp Med Biol. 2025;1468:171-175. doi: 10.1007/978-3-031-76550-6_28. Adv Exp Med Biol. 2025. PMID: 39930191 Review.
-
Phototactic preference and its genetic basis in the planulae of the colonial Hydrozoan Hydractinia symbiolongicarpus.bioRxiv [Preprint]. 2024 Apr 1:2024.03.28.585045. doi: 10.1101/2024.03.28.585045. bioRxiv. 2024. PMID: 38617216 Free PMC article. Preprint.
-
The USH3A causative gene clarin1 functions in Müller glia to maintain retinal photoreceptors.PLoS Genet. 2025 Mar 11;21(3):e1011205. doi: 10.1371/journal.pgen.1011205. eCollection 2025 Mar. PLoS Genet. 2025. PMID: 40067805 Free PMC article.
References
-
- Kimberling, W.J., Hildebrand, M.S., Shearer, A.E., Jensen, M.L., Halder, J.A., Trzupek, K., Cohn, E.S., Weleber, R.G., Stone, E.M. and Smith, R.J. (2010) Frequency of Usher syndrome in two pediatric populations: implications for genetic screening of deaf and hard of hearing children. Genet. Med., 12, 512–516. - PMC - PubMed
-
- Friedman, T.B., Schultz, J.M., Ahmed, Z.M., Tsilou, E.T. and Brewer, C.C. (2011) Usher syndrome: hearing loss with vision loss. Adv. Otorhinolaryngol., 70, 56–65. - PubMed
-
- May-Simera, H., Nagel-Wolfrum, K. and Wolfrum, U. (2017) Cilia – the sensory antennae in the eye. Prog. Retin. Eye Res., 60, 144–180. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

