ADAM17-mediated EGFR ligand shedding directs macrophage-promoted cancer cell invasion
- PMID: 35998057
- PMCID: PMC9675555
- DOI: 10.1172/jci.insight.155296
ADAM17-mediated EGFR ligand shedding directs macrophage-promoted cancer cell invasion
Abstract
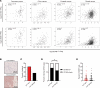
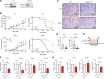
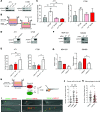
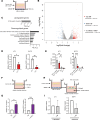
Macrophages in the tumor microenvironment have a substantial impact on tumor progression. Depending on the signaling environment in the tumor, macrophages can either support or constrain tumor progression. It is therefore of therapeutic interest to identify the tumor-derived factors that control macrophage education. With this aim, we correlated the expression of A Disintegrin and Metalloproteinase (ADAM) proteases, which are key mediators of cell-cell signaling, to the expression of protumorigenic macrophage markers in human cancer cohorts. We identified ADAM17, a sheddase upregulated in many cancer types, as a protein of interest. Depletion of ADAM17 in cancer cell lines reduced the expression of several protumorigenic markers in neighboring macrophages in vitro as well as in mouse models. Moreover, ADAM17-/- educated macrophages demonstrated a reduced ability to induce cancer cell invasion. Using mass spectrometry-based proteomics and ELISA, we identified heparin-binding EGF (HB-EGF) and amphiregulin, shed by ADAM17 in the cancer cells, as the implicated molecular mediators of macrophage education. Additionally, RNA-Seq and ELISA experiments revealed that ADAM17-dependent HB-EGF ligand release induced the expression and secretion of CXCL chemokines in macrophages, which in turn stimulated cancer cell invasion. In conclusion, we provide evidence that ADAM17 mediates a paracrine EGFR-ligand-chemokine feedback loop, whereby cancer cells hijack macrophages to promote tumor progression.
Keywords: Cancer; Macrophages; Oncology; Proteases.
Conflict of interest statement
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

