Multiancestry exome sequencing reveals INHBE mutations associated with favorable fat distribution and protection from diabetes
- PMID: 35999217
- PMCID: PMC9399235
- DOI: 10.1038/s41467-022-32398-7
Multiancestry exome sequencing reveals INHBE mutations associated with favorable fat distribution and protection from diabetes
Abstract
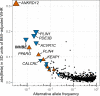
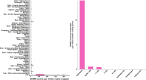
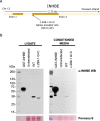
Body fat distribution is a major, heritable risk factor for cardiometabolic disease, independent of overall adiposity. Using exome-sequencing in 618,375 individuals (including 160,058 non-Europeans) from the UK, Sweden and Mexico, we identify 16 genes associated with fat distribution at exome-wide significance. We show 6-fold larger effect for fat-distribution associated rare coding variants compared with fine-mapped common alleles, enrichment for genes expressed in adipose tissue and causal genes for partial lipodystrophies, and evidence of sex-dimorphism. We describe an association with favorable fat distribution (p = 1.8 × 10-09), favorable metabolic profile and protection from type 2 diabetes (~28% lower odds; p = 0.004) for heterozygous protein-truncating mutations in INHBE, which encodes a circulating growth factor of the activin family, highly and specifically expressed in hepatocytes. Our results suggest that inhibin βE is a liver-expressed negative regulator of adipose storage whose blockade may be beneficial in fat distribution-associated metabolic disease.
© 2022. The Author(s).
Conflict of interest statement
Regeneron authors receive salary from and own options and/or stock of the company. G.D.Y is the Chief Scientific Officer and member of the Board of Directors at Regeneron Pharmaceuticals; A.J.M is an Executive Officer of Regeneron Pharmaceuticals. L.A.L., P.A., O.S., M.A.R.F., and A.B. are inventors on provisional patent applications (63/233,258 and 63/274,595), U.S. non-provisional applications (17/549,692, and 17/711,137), and PCT international application (PCT/US21/63150) submitted by RGC relating to
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- MC_EX_G0801669/MRC_/Medical Research Council/United Kingdom
- MC_UU_00017/3/MRC_/Medical Research Council/United Kingdom
- CRUK_/Cancer Research UK/United Kingdom
- CH/1996001/9454/BHF_/British Heart Foundation/United Kingdom
- MC_PC_12027/MRC_/Medical Research Council/United Kingdom
- MC_U137686854/MRC_/Medical Research Council/United Kingdom
- MC_UU_00017/2/MRC_/Medical Research Council/United Kingdom
- MC_PC_12029/MRC_/Medical Research Council/United Kingdom
- G0300622/MRC_/Medical Research Council/United Kingdom
- MC_PC_12028/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

