Differences in CD80 and CD86 transendocytosis reveal CD86 as a key target for CTLA-4 immune regulation
- PMID: 35999394
- PMCID: PMC9477731
- DOI: 10.1038/s41590-022-01289-w
Differences in CD80 and CD86 transendocytosis reveal CD86 as a key target for CTLA-4 immune regulation
Abstract
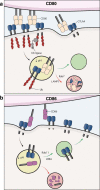
CD28 and CTLA-4 (CD152) play essential roles in regulating T cell immunity, balancing the activation and inhibition of T cell responses, respectively. Although both receptors share the same ligands, CD80 and CD86, the specific requirement for two distinct ligands remains obscure. In the present study, we demonstrate that, although CTLA-4 targets both CD80 and CD86 for destruction via transendocytosis, this process results in separate fates for CTLA-4 itself. In the presence of CD80, CTLA-4 remained ligand bound, and was ubiquitylated and trafficked via late endosomes and lysosomes. In contrast, in the presence of CD86, CTLA-4 detached in a pH-dependent manner and recycled back to the cell surface to permit further transendocytosis. Furthermore, we identified clinically relevant mutations that cause autoimmune disease, which selectively disrupted CD86 transendocytosis, by affecting either CTLA-4 recycling or CD86 binding. These observations provide a rationale for two distinct ligands and show that defects in CTLA-4-mediated transendocytosis of CD86 are associated with autoimmunity.
© 2022. The Author(s).
Conflict of interest statement
O.S.Q. is an employee of Celentix Ltd. The other authors declare no competing interests
Figures







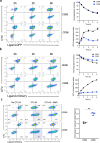


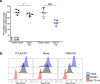
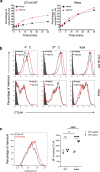
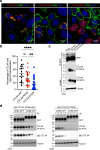
Comment in
-
Two CTLA-4 ligands, separate pHates.Nat Immunol. 2022 Sep;23(9):1297-1299. doi: 10.1038/s41590-022-01296-x. Nat Immunol. 2022. PMID: 35999395 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
- DH_/Department of Health/United Kingdom
- MC_UU_00008/4/MRC_/Medical Research Council/United Kingdom
- BB/H013598/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/H013598/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M009203/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 207547/Z/17/Z/WT_/Wellcome Trust/United Kingdom
- 110297/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- 102186/B/13/Z/WT_/Wellcome Trust/United Kingdom
- 204798/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- MR/N001435/1/MRC_/Medical Research Council/United Kingdom
- 21147/VAC_/Versus Arthritis/United Kingdom
- 204798/WT_/Wellcome Trust/United Kingdom
- BB/H013598/2/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

