Exploring actinomycetes natural products to identify potential multi-target inhibitors against Leishmania donovani
- PMID: 35999912
- PMCID: PMC9392678
- DOI: 10.1007/s13205-022-03304-1
Exploring actinomycetes natural products to identify potential multi-target inhibitors against Leishmania donovani
Abstract
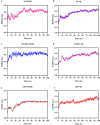
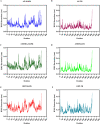
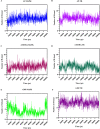
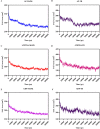
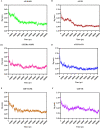
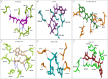
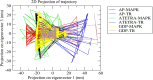
Visceral leishmaniasis (VL) is a neglected tropical disease that mainly affects the poor population of the Indian, African, and South American subcontinent. The increasing resistance to antimonial and miltefosine and frequent toxicity of amphotericin B drives an urgent need to develop an anti-leishmanial drug with excellent efficacy and safety profile. In this study, three sequential docking protocols (HTVS, SP, and XP) were performed to screen the secondary metabolites (n = 6519) from the actinomycetes source against five key proteins involved in the metabolic pathway of Leishmania donovani. Those proteins were adenine phosphoribosyltransferase (PDB ID: 1QB7), trypanothione reductase (PDB ID: 2JK6), N-myristoyl transferase (PDB ID: 2WUU), pteridine reductase (PDB ID: 2XOX), and MAP kinase (PDB ID: 4QNY). Although the binding energy of top ligands was predicted using the MM-GBSA module of the Schrödinger suite. SP and XP docking mode resulted in 55 multi-targeted ligands against L donovani. MM-GBSA analysis selected the top 18 ligands with good-binding affinity and the binding-free energy for four proteins, as mentioned earlier, when compared with the miltefosine, paromomycin, and a reference ligand selected for each target. Finally, molecular dynamics simulation, post-MD-binding-free energy (MM-PBSA), and principal component analysis (PCA) proposed three best ligands (Adenosine pentaphosphate, Atetra P, and GDP-4-keto-6-deoxymannose) qualifying the above screening parameters and confirmed as a potential drug candidate to fight against Leishmania donovani parasites.
Keywords: Actinomycetes; HTVS; Leishmania donovani; MM-PBSA; PCA; Secondary metabolites; Visceral leishmaniasis infection.
© King Abdulaziz City for Science and Technology 2022, Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Conflict of interest statement
Conflict of interestThe Authors have declared no competing interest.
Figures
Similar articles
-
Febrifugine analogues as Leishmania donovani trypanothione reductase inhibitors: binding energy analysis assisted by molecular docking, ADMET and molecular dynamics simulation.J Biomol Struct Dyn. 2017 Jan;35(1):141-158. doi: 10.1080/07391102.2015.1135298. Epub 2016 Apr 4. J Biomol Struct Dyn. 2017. PMID: 27043972
-
Developing imidazole analogues as potential inhibitor for Leishmania donovani trypanothione reductase: virtual screening, molecular docking, dynamics and ADMET approach.J Biomol Struct Dyn. 2015;33(12):2541-53. doi: 10.1080/07391102.2015.1085904. Epub 2015 Sep 28. J Biomol Struct Dyn. 2015. PMID: 26305585
-
Structure-based virtual screening, molecular docking, ADMET and molecular simulations to develop benzoxaborole analogs as potential inhibitor against Leishmania donovani trypanothione reductase.J Recept Signal Transduct Res. 2017 Feb;37(1):60-70. doi: 10.3109/10799893.2016.1171344. Epub 2016 May 5. J Recept Signal Transduct Res. 2017. PMID: 27147242
-
Natural products derived steroids as potential anti-leishmanial agents; disease prevalence, underlying mechanisms and future perspectives.Steroids. 2023 May;193:109196. doi: 10.1016/j.steroids.2023.109196. Epub 2023 Feb 9. Steroids. 2023. PMID: 36764565 Review.
-
Leishmaniasis: an update of current pharmacotherapy.Expert Opin Pharmacother. 2013 Jan;14(1):53-63. doi: 10.1517/14656566.2013.755515. Epub 2012 Dec 21. Expert Opin Pharmacother. 2013. PMID: 23256501 Review.
Cited by
-
Translational vaccinomics and structural filtration algorithm to device multiepitope vaccine for catastrophic monkeypox virus.Comput Biol Med. 2023 Feb;153:106497. doi: 10.1016/j.compbiomed.2022.106497. Epub 2022 Dec 30. Comput Biol Med. 2023. PMID: 36599210 Free PMC article.
-
Nanobody-peptide-conjugate (NPC) for passive immunotherapy against SARS-CoV-2 variants of concern (VoC): a prospective pan-coronavirus therapeutics.Mol Divers. 2023 Dec;27(6):2577-2603. doi: 10.1007/s11030-022-10570-x. Epub 2022 Nov 18. Mol Divers. 2023. PMID: 36400898 Free PMC article.
-
ACW-02 an Acridine Triazolidine Derivative Presents Antileishmanial Activity Mediated by DNA Interaction and Immunomodulation.Pharmaceuticals (Basel). 2023 Jan 29;16(2):204. doi: 10.3390/ph16020204. Pharmaceuticals (Basel). 2023. PMID: 37259353 Free PMC article.
-
Targeting with Structural Analogs of Natural Products the Purine Salvage Pathway in Leishmania (Leishmania) infantum by Computer-Aided Drug-Design Approaches.Trop Med Infect Dis. 2024 Feb 3;9(2):41. doi: 10.3390/tropicalmed9020041. Trop Med Infect Dis. 2024. PMID: 38393130 Free PMC article.
-
Identification of multi-targeting natural antiviral peptides to impede SARS-CoV-2 infection.Struct Chem. 2022 Dec 17:1-16. doi: 10.1007/s11224-022-02113-9. Online ahead of print. Struct Chem. 2022. PMID: 36570051 Free PMC article.
References
-
- Agarwal KC. Purines and myocardial protection. Boston, MA, US: Springer; 1996. Purines and regulation of platelet activation; pp. 409–418.
-
- Balasubramaniam DM, Williams DP (2020) WebGRO for Macromolecular Simulations.
LinkOut - more resources
Full Text Sources

