Computational modelling studies of some 1,3-thiazine derivatives as anti-influenza inhibitors targeting H1N1 neuraminidase via 2D-QSAR, 3D-QSAR, molecular docking, and ADMET predictions
- PMID: 36000144
- PMCID: PMC9389500
- DOI: 10.1186/s43088-022-00280-6
Computational modelling studies of some 1,3-thiazine derivatives as anti-influenza inhibitors targeting H1N1 neuraminidase via 2D-QSAR, 3D-QSAR, molecular docking, and ADMET predictions
Abstract
Background: Influenza virus disease remains one of the most contagious diseases that aided the deaths of many patients, especially in this COVID-19 pandemic era. Recent discoveries have shown that the high prevalence of influenza and SARS-CoV-2 coinfection can rapidly increase the death rate of patients. Hence, it became necessary to search for more potent inhibitors for influenza disease therapy. The present study utilized some computational modeling concepts such as 2D-QSAR, 3D-QSAR, molecular docking simulation, and ADMET predictions of some 1,3-thiazine derivatives as inhibitors of influenza neuraminidase (NA).
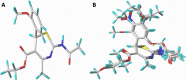
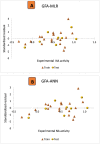
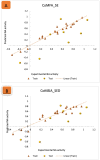
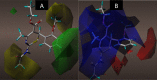
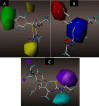
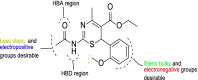
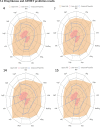
Results: The 2D-QSAR modeling results showed GFA-MLR ( = 0.9192, Q 2 = 0.8767, R 2 adj = 0.8991, RMSE = 0.0959, = 0.8943, = 0.7745) and GFA-ANN ( = 0.9227, Q 2 = 0.9212, RMSE = 0.0940, = 0.8831, = 0.7763) models with the computed descriptors as ATS7s, SpMax5_Bhv, nHBint6, and TDB9m for predicting the NA inhibitory activities of compounds which have passed the global criteria of accepting QSAR model. The 3D-QSAR modeling was carried out based on the comparative molecular field analysis (CoMFA) and comparative similarity indices analysis (CoMSIA). The CoMFA_ES ( = 0.9620, Q 2 = 0.643) and CoMSIA_SED ( = 0.8770, Q 2 = 0.702) models were found to also have good and reliable predicting ability. The compounds were also virtually screened based on their binding scores via molecular docking simulations with the active site of the NA (H1N1) target receptor which also confirms their resilient potency. Four potential lead compounds (4, 7, 14, and 15) with the relatively high inhibitory rate (> 50%) and docking (> - 6.3 kcal/mol) scores were identified as the possible lead candidates for in silico exploration of improved anti-influenza agents.
Conclusion: The drug-likeness and ADMET predictions of the lead compounds revealed non-violation of Lipinski's rule and good pharmacokinetic profiles as important guidelines for rational drug design. Hence, the outcome of this research set a course for the in silico design and exploration of novel NA inhibitors with improved potency.
Keywords: Binding score; Modeling; Neuraminidase; Receptor; Residual interaction.
© The Author(s) 2022.
Conflict of interest statement
Competing interestsThe authors declare that they have no competing interests.
Figures





References
-
- Akhtar Z, Islam MA, Aleem MA, Mah EMS, Ahmmed MK, Ghosh PK, et al. SARS-CoV-2 and influenza virus coinfection among patients with severe acute respiratory infection during the first wave of COVID-19 pandemic in Bangladesh: a hospital-based descriptive study. BMJ Open. 2021;11(11):e053768. doi: 10.1136/bmjopen-2021-053768. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
