Molecular and cellular architecture of the larval sensory organ in the cnidarian Nematostella vectensis
- PMID: 36000354
- PMCID: PMC9481973
- DOI: 10.1242/dev.200833
Molecular and cellular architecture of the larval sensory organ in the cnidarian Nematostella vectensis
Abstract
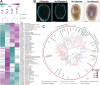
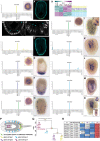
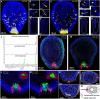
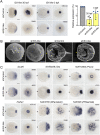
Cnidarians are the only non-bilaterian group to evolve ciliated larvae with an apical sensory organ, which is possibly homologous to the apical organs of bilaterian primary larvae. Here, we generated transcriptomes of the apical tissue in the sea anemone Nematostella vectensis and showed that it has a unique neuronal signature. By integrating previously published larval single-cell data with our apical transcriptomes, we discovered that the apical domain comprises a minimum of six distinct cell types. We show that the apical organ is compartmentalised into apical tuft cells (spot) and larval-specific neurons (ring). Finally, we identify ISX-like (NVE14554), a PRD class homeobox gene specifically expressed in apical tuft cells, as an FGF signalling-dependent transcription factor responsible for the formation of the apical tuft domain via repression of the neural ring fate in apical cells. With this study, we contribute a comparison of the molecular anatomy of apical organs, which must be carried out across phyla to determine whether this crucial larval structure evolved once or multiple times.
Keywords: Nematostella vectensis; Apical organ; Cilia; Cnidaria; Evolution; Neuron.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures




References
-
- Brauchle, M., Bilican, A., Eyer, C., Bailly, X., Martínez, P., Ladurner, P., Bruggmann, R. and Sprecher, S. G. (2018). Xenacoelomorpha survey reveals that all 11 animal homeobox gene classes were present in the first Bilaterians. Genome Biol. Evol. 10, 2205-2217. 10.1093/gbe/evy170 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

