Supplemental Oxygen Alters the Airway Microbiome in Cystic Fibrosis
- PMID: 36000724
- PMCID: PMC9601246
- DOI: 10.1128/msystems.00364-22
Supplemental Oxygen Alters the Airway Microbiome in Cystic Fibrosis
Abstract
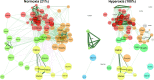
Features of the airway microbiome in persons with cystic fibrosis (pwCF) are correlated with disease progression. Microbes have traditionally been classified for their ability to tolerate oxygen. It is unknown whether supplemental oxygen, a common medical intervention, affects the airway microbiome of pwCF. We hypothesized that hyperoxia significantly impacts the pulmonary microbiome in cystic fibrosis. In this study, we cultured spontaneously expectorated sputum from pwCF in artificial sputum medium under 21%, 50%, and 100% oxygen conditions using a previously validated model system that recapitulates microbial community composition in uncultured sputum. Culture aliquots taken at 24, 48, and 72 h, along with uncultured sputum, underwent shotgun metagenomic sequencing with absolute abundance values obtained with the use of spike-in bacteria. Raw sequencing files were processed using the bioBakery pipeline to determine changes in taxonomy, predicted function, antimicrobial resistance genes, and mobile genetic elements. Hyperoxia reduced absolute microbial load, species richness, and diversity. Hyperoxia reduced absolute abundance of specific microbes, including facultative anaerobes such as Rothia and some Streptococcus species, with minimal impact on canonical CF pathogens such as Pseudomonas aeruginosa and Staphylococcus aureus. The effect size of hyperoxia on predicted functional pathways was stronger than that on taxonomy. Large changes in microbial cooccurrence networks were noted. Hyperoxia exposure perturbs airway microbial communities in a manner well tolerated by key pathogens. Supplemental oxygen use may enable the growth of lung pathogens and should be further studied in the clinical setting. IMPORTANCE The airway microbiome in persons with cystic fibrosis (pwCF) is correlated with lung function and disease severity. Supplemental oxygen use is common in more advanced CF, yet its role in perturbing airway microbial communities is unknown. By culturing sputum samples from pwCF under normal and elevated oxygen conditions, we found that increased oxygen led to reduced total numbers and diversity of microbes, with relative sparing of common CF pathogens such as Pseudomonas aeruginosa and Staphylococcus aureus. Supplemental oxygen use may enable the growth of lung pathogens and should be further studied in the clinical setting.
Keywords: cystic fibrosis; hyperoxia; lung; microbiome; oxygen; persons with cystic fibrosis; perturbation; pwCF.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
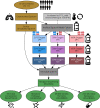

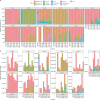
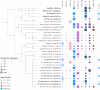
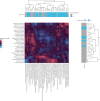
References
-
- Limoli DH, Yang J, Khansaheb MK, Helfman B, Peng L, Stecenko AA, Goldberg JB. 2016. Staphylococcus aureus and Pseudomonas aeruginosa co-infection is associated with cystic fibrosis-related diabetes and poor clinical outcomes. Eur J Clin Microbiol Infect Dis 35:947–953. doi:10.1007/s10096-016-2621-0. - DOI - PubMed
-
- Delhaes L, Monchy S, Fréalle E, Hubans C, Salleron J, Leroy S, Prevotat A, Wallet F, Wallaert B, Dei-Cas E, Sime-Ngando T, Chabé M, Viscogliosi E. 2012. The airway microbiota in cystic fibrosis: a complex fungal and bacterial community–implications for therapeutic management. PLoS One 7:e36313. doi:10.1371/journal.pone.0036313. - DOI - PMC - PubMed

