Orchestrated Biosynthesis of the Secondary Metabolite Cocktails Enables the Producing Fungus to Combat Diverse Bacteria
- PMID: 36000736
- PMCID: PMC9600275
- DOI: 10.1128/mbio.01800-22
Orchestrated Biosynthesis of the Secondary Metabolite Cocktails Enables the Producing Fungus to Combat Diverse Bacteria
Abstract
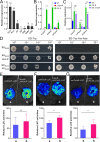
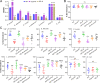
Fungal secondary metabolites with antibiotic activities can promote fungal adaptation to diverse environments. Besides the global regulator, individual biosynthetic gene clusters (BGCs) usually contain a pathway-specific transcription factor for the tight regulation of fungal secondary metabolism. Here, we report the chemical biology mediated by a supercluster containing three BGCs in the entomopathogenic fungus Metarhizium robertsii. These clusters are jointly controlled by an embedded transcription factor that orchestrates the collective production of four classes of chemicals: ustilaginoidin, indigotide, pseurotin, and hydroxyl-ovalicin. The ustilaginoidin BGC is implicated as a late-acquired cluster in Metarhizium to produce both the bis-naphtho-γ-pyrones and the monomeric naphtho-γ-pyrone glycosides (i.e., indigotides). We found that the biosynthesis of indigotides additionally requires the functions of paired methylglucosylation genes located outside the supercluster. The pseurotin/ovalicin BGCs are blended and mesosyntenically conserved to the intertwined pseurotin/fumagillin BGCs of Aspergillus fumigatus. However, the former have lost a few genes, including a polyketide synthase gene responsible for the production of a pentaene chain used for assembly with ovalicin to form fumagillin, as observed in A. fumigatus. The collective production of chemical cocktails by this supercluster was dispensable for fungal virulence against insects and could enable the fungus to combat different bacteria better than the metabolite(s) produced by an individual BGC could. Thus, our results unveil a novel strategy employed by fungi to manage chemical ecology against diverse bacteria. IMPORTANCE Fungal chemical ecology is largely mediated by the metabolite(s) produced by individual biosynthetic gene clusters (BGCs) with antibiotic activities. We report a supercluster containing three BGCs that are jointly controlled by an embedded master regulator in the insect pathogen Metarhizium robertsii. Four classes of chemicals, namely, ustilaginoidin, indigotide, pseurotin, and hydroxyl-ovalicin, are collectively produced by these three BGCs along with the contributions of tailoring enzyme genes located outside the supercluster. The production of these metabolites is not required for the fungal infection of insect hosts, but it benefits the fungus to combat diverse bacteria. The findings reveal and advocate a "the-more-the-better" strategy employed by fungi to manage effective adaptations to diverse environments.
Keywords: Metarhizium; chemical ecology; master regulator; secondary metabolism; supercluster.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Inductive Production of the Iron-Chelating 2-Pyridones Benefits the Producing Fungus To Compete for Diverse Niches.mBio. 2021 Dec 21;12(6):e0327921. doi: 10.1128/mbio.03279-21. Epub 2021 Dec 14. mBio. 2021. PMID: 34903054 Free PMC article.
-
Secondary metabolite gene clusters in the entomopathogen fungus Metarhizium anisopliae: genome identification and patterns of expression in a cuticle infection model.BMC Genomics. 2016 Oct 25;17(Suppl 8):736. doi: 10.1186/s12864-016-3067-6. BMC Genomics. 2016. PMID: 27801295 Free PMC article.
-
Transcription Factor Repurposing Offers Insights into Evolution of Biosynthetic Gene Cluster Regulation.mBio. 2021 Aug 31;12(4):e0139921. doi: 10.1128/mBio.01399-21. Epub 2021 Jul 20. mBio. 2021. PMID: 34281384 Free PMC article.
-
Complexity of fungal polyketide biosynthesis and function.Mol Microbiol. 2024 Jan;121(1):18-25. doi: 10.1111/mmi.15192. Epub 2023 Nov 13. Mol Microbiol. 2024. PMID: 37961029 Review.
-
Presence, Mode of Action, and Application of Pathway Specific Transcription Factors in Aspergillus Biosynthetic Gene Clusters.Int J Mol Sci. 2021 Aug 13;22(16):8709. doi: 10.3390/ijms22168709. Int J Mol Sci. 2021. PMID: 34445420 Free PMC article. Review.
Cited by
-
Biosynthetic gene cluster synteny: Orthologous polyketide synthases in Hypogymnia physodes, Hypogymnia tubulosa, and Parmelia sulcata.Microbiologyopen. 2023 Oct;12(5):e1386. doi: 10.1002/mbo3.1386. Microbiologyopen. 2023. PMID: 37877655 Free PMC article.
-
Engineered fungus containing a caterpillar gene kills insects rapidly by disrupting their ecto- and endo-microbiomes.Commun Biol. 2024 Aug 7;7(1):955. doi: 10.1038/s42003-024-06670-z. Commun Biol. 2024. PMID: 39112633 Free PMC article.
-
Analyzing the Challenges and Opportunities Associated With Harnessing New Antibiotics From the Fungal Microbiome.Microbiologyopen. 2025 Aug;14(4):e70034. doi: 10.1002/mbo3.70034. Microbiologyopen. 2025. PMID: 40698515 Free PMC article. Review.
-
Comparative genomics of Metarhizium brunneum strains V275 and ARSEF 4556: unraveling intraspecies diversity.G3 (Bethesda). 2024 Oct 7;14(10):jkae190. doi: 10.1093/g3journal/jkae190. G3 (Bethesda). 2024. PMID: 39210673 Free PMC article.
-
Towards a Rational Basis for the Selection of Probiotics to Improve Silkworm Health and Performance.Insects. 2025 Feb 4;16(2):162. doi: 10.3390/insects16020162. Insects. 2025. PMID: 40003792 Free PMC article. Review.
References
-
- Bergmann S, Funk AN, Scherlach K, Schroeckh V, Shelest E, Horn U, Hertweck C, Brakhage AA. 2010. Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic nonribosomal peptide synthetase gene cluster. Appl Environ Microbiol 76:8143–8149. doi:10.1128/AEM.00683-10. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

