The Omicron Variant BA.1.1 Presents a Lower Pathogenicity than B.1 D614G and Delta Variants in a Feline Model of SARS-CoV-2 Infection
- PMID: 36000850
- PMCID: PMC9472624
- DOI: 10.1128/jvi.00961-22
The Omicron Variant BA.1.1 Presents a Lower Pathogenicity than B.1 D614G and Delta Variants in a Feline Model of SARS-CoV-2 Infection
Abstract
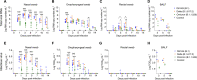
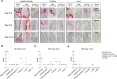
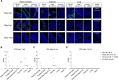
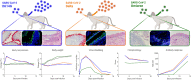
Omicron (B.1.1.529) is the most recent SARS-CoV-2 variant of concern, which emerged in late 2021 and rapidly achieved global predominance by early 2022. In this study, we compared the infection dynamics, tissue tropism, and pathogenesis and pathogenicity of SARS-CoV-2 D614G (B.1), Delta (B.1.617.2), and Omicron BA.1.1 (B.1.1.529) variants in a highly susceptible feline model of infection. Although D614G- and Delta-inoculated cats became lethargic and showed increased body temperatures between days 1 and 3 postinfection (pi), Omicron-inoculated cats remained subclinical and, similar to control animals, gained weight throughout the 14-day experimental period. Intranasal inoculation of cats with D614G- and the Delta variants resulted in high infectious virus shedding in nasal secretions (up to 6.3 log10 TCID50.Ml-1), whereas strikingly lower level of viruses shedding (<3.1 log10 TCID50.Ml-1) was observed in Omicron-inoculated animals. In addition, tissue distribution of the Omicron variant was markedly reduced in comparison to the D614G and Delta variants, as evidenced by lower in situ viral RNA detection, in situ viral immunofluorescence staining, and viral loads in tissues on days 3, 5, and 14 pi. Nasal turbinate, trachea, and lung were the main-but not the only-sites of replication for all three viral variants. However, only scarce virus staining and lower viral titers suggest lower levels of viral replication in tissues from Omicron-infected animals. Notably, while D614G- and Delta-inoculated cats presented pneumonia, histologic examination of the lungs from Omicron-infected cats revealed mild to modest inflammation. Together, these results demonstrate that the Omicron variant BA.1.1 is less pathogenic than D614G and Delta variants in a highly susceptible feline model. IMPORTANCE The SARS-CoV-2 Omicron (B.1.1.529) variant of concern emerged in South Africa late in 2021 and rapidly spread across the world causing a significant increase in the number of infections. Importantly, this variant was also associated with an increased risk of reinfections. However, the number of hospitalizations and deaths due to COVID-19 did not follow the same trends. These early observations suggested effective protection conferred by immunizations and/or overall lower virulence of the highly mutated variant virus. In this study we present novel evidence demonstrating that the Omicron BA.1.1 variant of concern presents a lower pathogenicity when compared to D614G- or Delta variants in cats. Clinical, virological, and pathological evaluations revealed lower disease severity, viral replication, and lung pathology in Omicron-infected cats when compared with D614G and Delta variant inoculated animals, confirming that Omicron BA.1.1 is less pathogenic in a highly susceptible feline model of infection.
Keywords: COVID-19; Delta; Omicron; SARS-CoV-2; cats; pathogenesis; pathogenicity; variant of concern.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Update of
-
The Omicron variant BA.1.1 presents a lower pathogenicity than B.1 D614G and Delta variants in a feline model of SARS-CoV-2 infection.bioRxiv [Preprint]. 2022 Jun 16:2022.06.15.496220. doi: 10.1101/2022.06.15.496220. bioRxiv. 2022. Update in: J Virol. 2022 Sep 14;96(17):e0096122. doi: 10.1128/jvi.00961-22. PMID: 35734088 Free PMC article. Updated. Preprint.
Similar articles
-
The Omicron variant BA.1.1 presents a lower pathogenicity than B.1 D614G and Delta variants in a feline model of SARS-CoV-2 infection.bioRxiv [Preprint]. 2022 Jun 16:2022.06.15.496220. doi: 10.1101/2022.06.15.496220. bioRxiv. 2022. Update in: J Virol. 2022 Sep 14;96(17):e0096122. doi: 10.1128/jvi.00961-22. PMID: 35734088 Free PMC article. Updated. Preprint.
-
The SARS-CoV-2 Spike is a virulence determinant and plays a major role on the attenuated phenotype of Omicron virus in a feline model of infection.J Virol. 2024 Mar 19;98(3):e0190223. doi: 10.1128/jvi.01902-23. Epub 2024 Feb 29. J Virol. 2024. PMID: 38421180 Free PMC article.
-
Comparative analysis of neutrophil dynamics and disease in SARS-CoV-2 Delta and Omicron variants utilizing an in vivo feline model for COVID-19.Front Immunol. 2025 May 22;16:1547918. doi: 10.3389/fimmu.2025.1547918. eCollection 2025. Front Immunol. 2025. PMID: 40475786 Free PMC article.
-
The Biological Functions and Clinical Significance of SARS-CoV-2 Variants of Corcern.Front Med (Lausanne). 2022 May 20;9:849217. doi: 10.3389/fmed.2022.849217. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35669924 Free PMC article. Review.
-
Evolution of the SARS-CoV-2 omicron variants BA.1 to BA.5: Implications for immune escape and transmission.Rev Med Virol. 2022 Sep;32(5):e2381. doi: 10.1002/rmv.2381. Epub 2022 Jul 20. Rev Med Virol. 2022. PMID: 35856385 Free PMC article. Review.
Cited by
-
A linear SARS-CoV-2 DNA vaccine candidate reduces virus shedding in ferrets.Arch Virol. 2023 Mar 29;168(4):124. doi: 10.1007/s00705-023-05746-1. Arch Virol. 2023. PMID: 36988739 Free PMC article.
-
Changes in Clinical Features and Severity of COVID-19 with the Emergence of Omicron Variants: A Shift Towards a Common Disease.Infect Drug Resist. 2024 Dec 18;17:5595-5603. doi: 10.2147/IDR.S492816. eCollection 2024. Infect Drug Resist. 2024. PMID: 39711829 Free PMC article.
-
Evaluating the distribution and clustering of SARS-CoV-2 antibodies in dogs across the United States of America.Sci Rep. 2025 Jul 1;15(1):21758. doi: 10.1038/s41598-025-06730-2. Sci Rep. 2025. PMID: 40593062 Free PMC article.
-
Analysis of SARS-CoV-2 genome evolutionary patterns.Microbiol Spectr. 2024 Feb 6;12(2):e0265423. doi: 10.1128/spectrum.02654-23. Epub 2024 Jan 10. Microbiol Spectr. 2024. PMID: 38197644 Free PMC article.
-
Assessing the Potential Role of Cats (Felis catus) as Generators of Relevant SARS-CoV-2 Lineages during the Pandemic.Pathogens. 2023 Nov 16;12(11):1361. doi: 10.3390/pathogens12111361. Pathogens. 2023. PMID: 38003825 Free PMC article.
References
-
- Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich AM, Neuman BW, Penzar D, Perlman S, Poon LLM, Samborskiy DV, Sidorov IA, Sola I, Ziebuhr J. 2020. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5:536–544. https://www.nature.com/articles/s41564-020-0695-z. - PMC - PubMed
-
- Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, Angyal A, Brown RL, Carrilero L, Green LR, Groves DC, Johnson KJ, Keeley AJ, Lindsey BB, Parsons PJ, Raza M, Rowland-Jones S, Smith N, Tucker RM, Wang D, Wyles MD, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC. 2020. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 182:812–827.e19. 10.1016/j.cell.2020.06.043. - DOI - PMC - PubMed
-
- Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, Hinsley WR, Laydon DJ, Dabrera G, O’Toole A, Amato R, Manon R-C, Harrison I, Jackson B, Ariani Cristina V, Boyd O, Loman NJ, McCrone JT, Gonçalves S, Jorgensen D, Myers R, Hill V, Jackson David K, Gaythorpe K, Groves N, Sillitoe J, Kwiatkowski DP, Flaxman S, Ratmann O, Bhatt S, Hopkins S, Gandy A, Rambaut A, Ferguson NM, The COVID-19 Genomics UK Consortium (COG-UK) . 2021. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 593:266–269. https://www.nature.com/articles/s41586-021-03470-x. - PubMed
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

