Predicting cross-tissue hormone-gene relations using balanced word embeddings
- PMID: 36000859
- PMCID: PMC9563690
- DOI: 10.1093/bioinformatics/btac578
Predicting cross-tissue hormone-gene relations using balanced word embeddings
Abstract
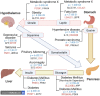
Motivation: Inter-organ/inter-tissue communication is central to multi-cellular organisms including humans, and mapping inter-tissue interactions can advance system-level whole-body modeling efforts. Large volumes of biomedical literature have fostered studies that map within-tissue or tissue-agnostic interactions, but literature-mining studies that infer inter-tissue relations, such as between hormones and genes are solely missing.
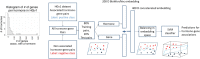
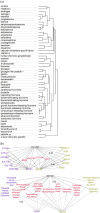
Results: We present a first study to predict from biomedical literature the hormone-gene associations mediating inter-tissue signaling in the human body. Our BioEmbedS* models use neural network-based Biomedical word Embeddings with a Support Vector Machine classifier to predict if a hormone-gene pair is associated or not, and whether an associated gene is involved in the hormone's production or response. Model training relies on our unified dataset Hormone-Gene version 1 of ground-truth associations between genes and endocrine hormones, which we compiled and carefully balanced in the embedded space to handle data disparities, such as between poorly- versus well-studied hormones. Our BioEmbedS model recapitulates known gene mediators of tissue-tissue signaling with 70.4% accuracy; predicts novel inter-tissue communication genes in humans, which are enriched for hormone-related disorders; and generalizes well to mouse, thereby holding promise for its extension to other multi-cellular organisms as well.
Availability and implementation: Freely available at https://cross-tissue-signaling.herokuapp.com are our model predictions & datasets; https://github.com/BIRDSgroup/BioEmbedS has all relevant code.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2022. Published by Oxford University Press.
Figures



References
-
- Argilés J.M. et al. (2018) Inter-tissue communication in cancer cachexia. Nat. Rev. Endocrinol., 15, 9–20. - PubMed
-
- Bojanowski P. et al. (2017) Enriching word vectors with subword information. TACL, 5, 135–146.
-
- Braschi B. et al. (2019) Genenames.org: the HGNC and VGNC resources in 2019. Nucleic Acids Res., 47, D786–D792. - PMC - PubMed

