Auxin and abscisic acid antagonistically regulate ascorbic acid production via the SlMAPK8-SlARF4-SlMYB11 module in tomato
- PMID: 36000899
- PMCID: PMC9614483
- DOI: 10.1093/plcell/koac262
Auxin and abscisic acid antagonistically regulate ascorbic acid production via the SlMAPK8-SlARF4-SlMYB11 module in tomato
Abstract
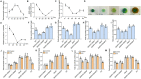
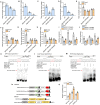
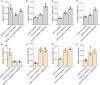
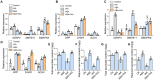
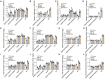
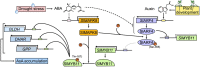
Ascorbic acid (AsA) is a multifunctional phytonutrient that is essential for the human diet as well as plant development. While much is known about AsA biosynthesis in plants, how this process is regulated in tomato (Solanum lycopersicum) fruits remains unclear. Here, we found that auxin treatment inhibited AsA accumulation in the leaves and pericarps of tomato. The auxin response factor gene SlARF4 is induced by auxin to mediate auxin-induced inhibition of AsA accumulation. Specifically, SlARF4 transcriptionally inhibits the transcription factor gene SlMYB11, thereby modulating AsA accumulation by regulating the transcription of the AsA biosynthesis genes l-galactose-1-phosphate phosphatase, l-galactono-1,4-lactone dehydrogenase, and dehydroascorbate. By contrast, abscisic acid (ABA) treatment increased AsA accumulation in tomato under drought stress. ABA induced the expression of the mitogen-activated protein kinase gene SlMAPK8. We demonstrate that SlMAPK8 phosphorylates SlARF4 and inhibits its transcriptional activity, whereas SlMAPK8 phosphorylates SlMYB11 and activates its transcriptional activity. SlMAPK8 functions in ABA-induced AsA accumulation and drought stress tolerance. Moreover, ABA antagonizes the effects of auxin on AsA biosynthesis. Therefore, auxin- and ABA-induced regulation of AsA accumulation is mediated by the SlMAPK8-SlARF4-SlMYB11 module in tomato during fruit development and drought stress responses, shedding light on the roles of phytohormones in regulating AsA accumulation to mediate stress tolerance.
© American Society of Plant Biologists 2022. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures






Similar articles
-
SlMYB99-mediated auxin and abscisic acid antagonistically regulate ascorbic acids biosynthesis in tomato.New Phytol. 2023 Aug;239(3):949-963. doi: 10.1111/nph.18988. Epub 2023 May 29. New Phytol. 2023. PMID: 37247338
-
Knockout of Auxin Response Factor SlARF4 Improves Tomato Resistance to Water Deficit.Int J Mol Sci. 2021 Mar 25;22(7):3347. doi: 10.3390/ijms22073347. Int J Mol Sci. 2021. PMID: 33805879 Free PMC article.
-
SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development.Plant Physiol. 2013 Mar;161(3):1362-74. doi: 10.1104/pp.113.213843. Epub 2013 Jan 22. Plant Physiol. 2013. PMID: 23341361 Free PMC article.
-
Roles of abscisic acid and auxin in plants during drought: A molecular point of view.Plant Physiol Biochem. 2023 Nov;204:108129. doi: 10.1016/j.plaphy.2023.108129. Epub 2023 Oct 20. Plant Physiol Biochem. 2023. PMID: 37897894 Review.
-
Multiple Physiological and Biochemical Functions of Ascorbic Acid in Plant Growth, Development, and Abiotic Stress Response.Int J Mol Sci. 2024 Feb 2;25(3):1832. doi: 10.3390/ijms25031832. Int J Mol Sci. 2024. PMID: 38339111 Free PMC article. Review.
Cited by
-
Fruit crops combating drought: Physiological responses and regulatory pathways.Plant Physiol. 2023 Jul 3;192(3):1768-1784. doi: 10.1093/plphys/kiad202. Plant Physiol. 2023. PMID: 37002821 Free PMC article. Review.
-
Identification of Transcription Factors of Santalene Synthase Gene Promoters and SaSSY Cis-Elements through Yeast One-Hybrid Screening in Santalum album L.Plants (Basel). 2024 Jul 8;13(13):1882. doi: 10.3390/plants13131882. Plants (Basel). 2024. PMID: 38999721 Free PMC article.
-
Identifying Candidate Genes Related to the Nutritional Components of Soybean (Glycine max) Sprouts Based on the Transcriptome and Co-Expression Network.Genes (Basel). 2025 Jun 6;16(6):692. doi: 10.3390/genes16060692. Genes (Basel). 2025. PMID: 40565584 Free PMC article.
-
ETHYLENE-INSENSITIVE 3-LIKE 2 regulates β-carotene and ascorbic acid accumulation in tomatoes during ripening.Plant Physiol. 2023 Jul 3;192(3):2067-2080. doi: 10.1093/plphys/kiad151. Plant Physiol. 2023. PMID: 36891812 Free PMC article.
-
AUXIN RESPONSE FACTOR 2 mediates repression of strawberry receptacle ripening via auxin-ABA interplay.Plant Physiol. 2024 Dec 2;196(4):2638-2653. doi: 10.1093/plphys/kiae510. Plant Physiol. 2024. PMID: 39405162 Free PMC article.
References
-
- Bouzroud S, Gasparini K, Hu G, Barbosa MAM, Rosa BL, Fahr M, Bendaou N, Bouzayen M, Zsögön A, Smouni A, et al. (2020) Down regulation and loss of auxin response factor 4 function using CRISPR/Cas9 alters plant growth, stomatal function and improves tomato tolerance to salinity and osmotic stress. Genes 11: 272 - PMC - PubMed
-
- Bulley S, Laing W (2016) The regulation of ascorbate biosynthesis. Curr Opin Plant Biol 33: 15–22 - PubMed
-
- Chen L, Sun H, Wang FJ, Yue DD, Shen XK, Sun WN, Zhang XL, Yang XY (2020) Genome-wide identification of MAPK cascade genes reveals the GhMAP3K14–GhMKK11–GhMPK31 pathway is involved in the drought response in cotton. Plant Mol Biol 103: 211–223 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

