Comparative Genomics Analysis and Outer Membrane Vesicle-Mediated Horizontal Antibiotic-Resistance Gene Transfer in Avibacterium paragallinarum
- PMID: 36000914
- PMCID: PMC9603892
- DOI: 10.1128/spectrum.01379-22
Comparative Genomics Analysis and Outer Membrane Vesicle-Mediated Horizontal Antibiotic-Resistance Gene Transfer in Avibacterium paragallinarum
Abstract
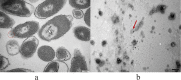
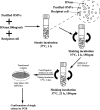
Avibacterium paragallinarum is the etiological agent of infectious coryza, an acute respiratory disease of chickens that is globally distributed and causes serious economic losses for chicken production. A. paragallinarum is a Gram-negative bacterium that releases outer membrane vesicles (OMVs). In this study, a comparative genomic analysis of A. paragallinarum isolate P4chr1 and its OMVs was carried out, and the ability to transfer antibiotic resistance genes (ARGs) via the OMVs was studied. Sequencing and data analyses demonstrated that the genomic size of A. paragallinarum P4chr1 was approximately 2.77 Mb with a 25 kb tolerance island that covered six types of antibiotics and 11 ARGs. The genomic size of its OMVs was approximately 2.69 Mb, covering 97% of the genomic length and almost all the gene sequences of P4chr1. Purified and DNase-treated A. paragallinarum P4chr1 OMVs were cocultured with the antibiotic-sensitive A. paragallinarum Modesto strain on an antibiotic (chloramphenicol, erythromycin, tetracycline, or streptomycin)-containing plate, and the corresponding ARGs were detected in the colonies grown on the plates. However, using an antimicrobial susceptibility test, we found that ARGs delivered by OMVs were not persistent but only appeared transiently on the antibiotic-containing plates. Antibiotic resistance and ARGs were lost by the second bacterial passage. IMPORTANCE The functions and roles of OMVs on ARG and virulent gene transfer and dissemination have been reported in numerous Gram-negative bacteria. However, the role of OMVs in mediating antibiotic resistance in A. paragallinarum has not been reported. This study is the first report to compare the genomic characteristics of OMVs with its parent A. paragallinarum strain and to study A. paragallinarum ARG transfer via OMVs. This work has provided useful data for further studies focusing on nonplasmid ARG transfer mediated by A. paragallinarum OMVs.
Keywords: A. paragallinarum; antibiotic resistance gene; horizontal gene transfer; outer membrane vesicles; whole genome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Prevalence and genomic-based antimicrobial resistance analysis of Avibacterium paragallinarum isolates in Guangdong Province, China.Poult Sci. 2024 Jun;103(6):103751. doi: 10.1016/j.psj.2024.103751. Epub 2024 Apr 8. Poult Sci. 2024. PMID: 38652951 Free PMC article.
-
Isolation, molecular detection, and sequence analysis of Avibacterium paragallinarum from suspected cases of infectious coryza infected chickens from different areas of Ethiopia, 2022-2024.BMC Microbiol. 2025 Mar 15;25(1):141. doi: 10.1186/s12866-025-03862-3. BMC Microbiol. 2025. PMID: 40087555 Free PMC article.
-
Antimicrobial susceptibility of Avibacterium paragallinarum isolates from outbreaks of infectious coryza in Dutch commercial poultry flocks, 2008-2017.Vet Microbiol. 2018 Apr;217:135-143. doi: 10.1016/j.vetmic.2018.03.008. Epub 2018 Mar 14. Vet Microbiol. 2018. PMID: 29615246
-
Bacterial Outer Membrane Vesicles as Antibiotic Delivery Vehicles.Front Immunol. 2021 Sep 20;12:733064. doi: 10.3389/fimmu.2021.733064. eCollection 2021. Front Immunol. 2021. PMID: 34616401 Free PMC article. Review.
-
Bacterial outer membrane vesicles as potential biological nanomaterials for antibacterial therapy.Acta Biomater. 2022 Mar 1;140:102-115. doi: 10.1016/j.actbio.2021.12.005. Epub 2021 Dec 9. Acta Biomater. 2022. PMID: 34896632 Review.
Cited by
-
Recommendation of a standardized broth microdilution method for antimicrobial susceptibility testing of Avibacterium paragallinarum and resistance monitoring.J Clin Microbiol. 2024 Mar 13;62(3):e0101123. doi: 10.1128/jcm.01011-23. Epub 2024 Feb 16. J Clin Microbiol. 2024. PMID: 38363142 Free PMC article.
-
The characterization of outer membrane vesicles (OMVs) and their role in mediating antibiotic-resistance gene transfer through natural transformation in Riemerella anatipestifer.Poult Sci. 2025 Feb;104(2):104730. doi: 10.1016/j.psj.2024.104730. Epub 2024 Dec 26. Poult Sci. 2025. PMID: 39729729 Free PMC article.
-
Development and Validation of PCR Diagnostic Assays for Detection of Avibacterium paragallinarum and Ornithobacterium rhinotracheale.Vet Sci. 2023 Dec 22;11(1):7. doi: 10.3390/vetsci11010007. Vet Sci. 2023. PMID: 38250913 Free PMC article.
-
Microbial extracellular vesicles contribute to antimicrobial resistance.PLoS Pathog. 2024 May 2;20(5):e1012143. doi: 10.1371/journal.ppat.1012143. eCollection 2024 May. PLoS Pathog. 2024. PMID: 38696356 Free PMC article. Review.
-
The effect of site-specific recombinases XerCD on the removal of over-replicated chromosomal DNA through outer membrane vesicles in bacteria.Microbiol Spectr. 2024 Mar 5;12(3):e0234323. doi: 10.1128/spectrum.02343-23. Epub 2024 Feb 13. Microbiol Spectr. 2024. PMID: 38349173 Free PMC article.
References
-
- Blackall PJ, Soriano-Vargas VE. 2020. Infectious coryza and related bacterial infections, p 890–906. In Swayne DE, Boulianne M, Logue CM, McDougald LR, Nair V, Suarez DL, de Wit S, Grimes T, Johnson D, Kromm M, Prajitno TY, Rubinoff I, Zavala G. (ed), Diseases of poultry. John Wiley & Sons, New York, NY. doi:10.1002/9781119371199.ch20. - DOI
-
- Heuvelink A, Wiegel J, Kehrenberg C, Dijkman R, Soriano VE, Feberwee A. 2018. Antimicrobial susceptibility of Avibacterium paragallinarum isolates from outbreaks of infectious coryza in Dutch commercial poultry flocks, 2008–2017. Vet Microbiol 217:135–143. doi:10.1016/j.vetmic.2018.03.008. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

